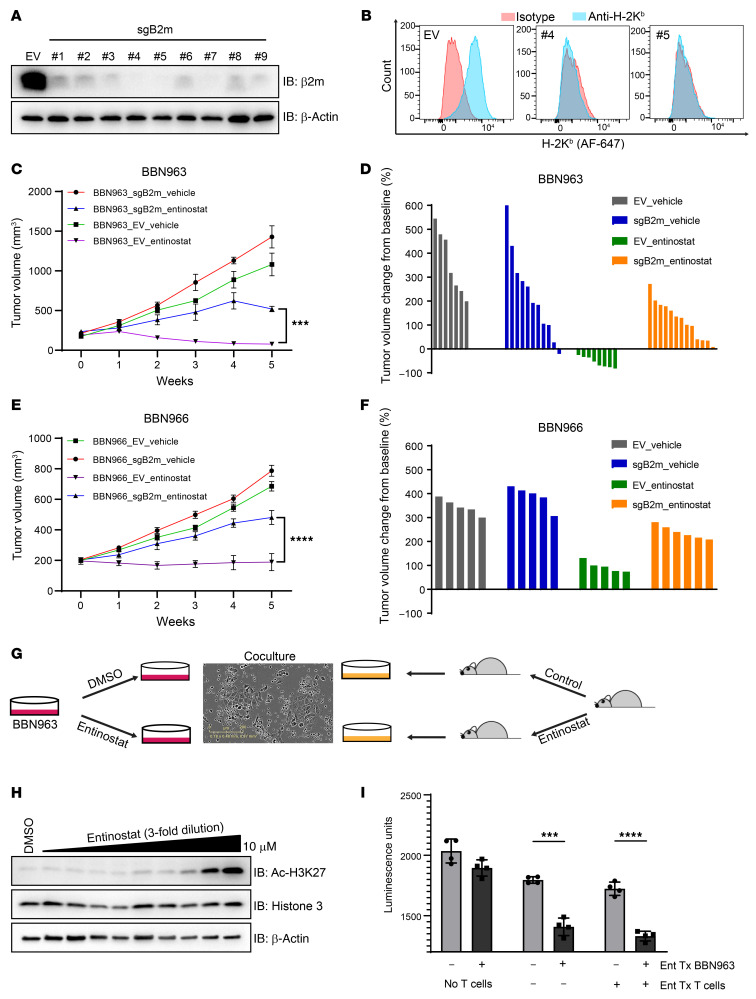
Figure 5. B2m CRISPR partially rescues the antitumor effect of entinostat.
(A) Immunoblot of BBN963 cells with B2m knockout using 9 different individual sgRNA constructs. (B) Flow cytometry graph of H-2Kb (class 1 MHC) cell-surface expression in B2m-knockout BBN963 cells in A with sgRNA no. 4 and no. 5. (C) Average volume of BBN963_EV (empty vector) and BBN963_sgB2m (B2m knockout) tumors in response to entinostat (12 mpk) in C57BL/6 mice. n = 7–13 mice per group. Significance was calculated using 2-way ANOVA followed by Tukey’s multiple-comparison test. (D) Waterfall plot of individual tumor volumes in C at end points relative to pretreatment baseline. End points were tumor burden and ulceration. (E) Average volume of BBN966_EV (empty vector) and BBN966_sgB2m (B2m knockout) tumors in response to entinostat (12 mpk) in C57BL/6 mice. n = 5–6 mice per group. Significance was calculated using 2-way ANOVA followed by Tukey’s multiple-comparison test. (F) Waterfall plot of individual tumor volumes in E at end points relative to pretreatment baseline. End points were tumor burden and ulceration (G) Schematic of how the in vitro T cell killing assays were set up. (H) Immunoblots of BBN963 cells treated with various concentrations of entinostat for 24 hours and blotted for the indicated antibodies. (I) Bar graph showing the result of T cell killing assays. CellTiter-Glo was used to quantify viable BBN963 cells at the end of the 72-hour coculture. Significance was calculated using t test. Data are represented as mean ± SD. ***P < 0.001; ****P < 0.0001.