Figure 4.
TCR clonotyping and single-cell RNA sequencing of T cells
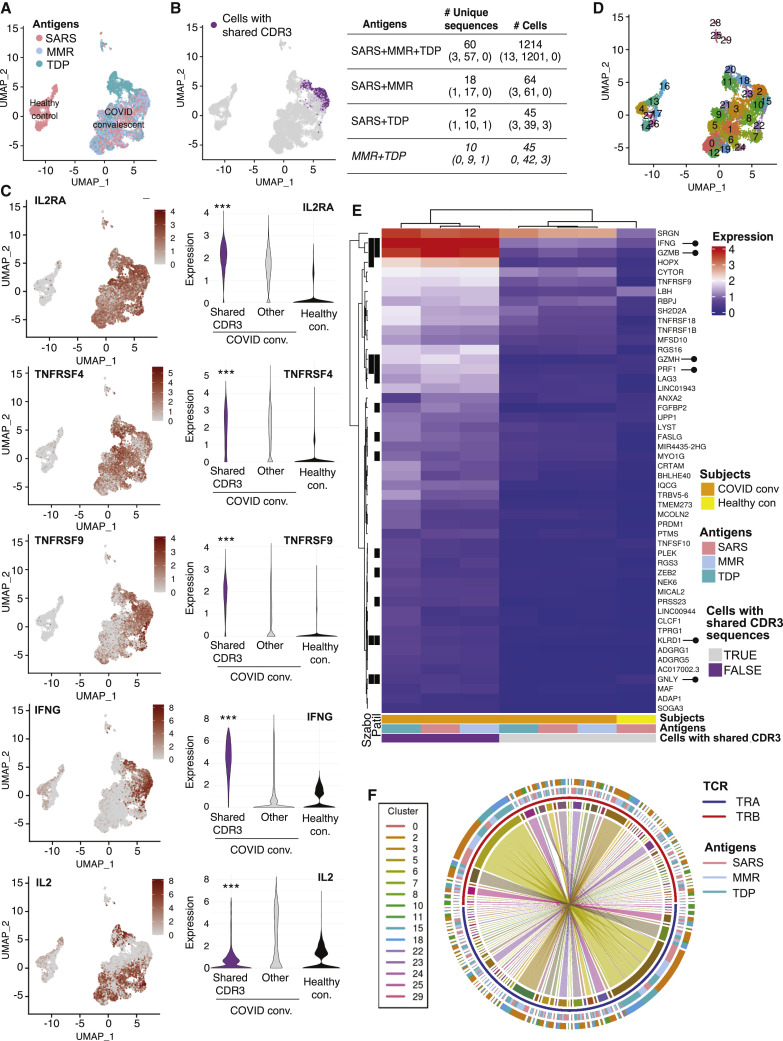
(A) Uniform manifold approximation and projection (UMAP) plots of single-cell gene expression data of T cells from uninfected (healthy) control and COVID-19-convalescent donors colored by the antigen to which they were exposed.
(B) UMAP plot highlighting cells with shared CDR3 sequences (left). Number of clonotypes and cells across SARS-CoV-2 (SARS), MMR, and Tdap (TDP) antigens overall and by each COVID-19-convalescent donor (in parentheses) is given.
(C) UMAP plot of single-cell gene expression data (left) and violin plots (right) showing the distribution of expression for indicated genes. Adjusted p values for differential expression were obtained from Wilcoxon rank-sum tests, as implemented in the FindMarkers function in the Seurat R package. ∗∗∗p < 0.001 compared with other groups.
(D) UMAP plot displaying the clustering scheme.
(E) Heatmap of mean expression of top 50 marker genes for cells with shared CDR3 sequences by COVID-convalescent status and antigen. The black bars on the left-hand side indicate that the gene was identified as a TEMRA marker in Patil et al.46 (column labeled “Patil”) or in Szabo et al.47 (column labeled “Szabo”). Rows and columns were grouped by overall expression pattern using hierarchical clustering. Lollipops (right) highlight cytotoxic T cell effector molecule genes.
(F) Circos plot of sequences with shared CDR3 sequences. Each sector on the innermost track corresponds to an α chain (TRA, blue) or β chain (TRB, red). The width of a sector is proportional to the number of times the α or β chain sequence occurs. An arc between an α and β chain indicates these sequences are combined in a single CDR3 sequence. The antigen is shown in the third track. The Seurat cluster is shown in the fourth track; not all clusters have cells with shared CDR3 sequences, so fewer than 30 clusters are shown. The preponderance of straight lines spanning across the plot shows that in nearly all cases, a given α chain is combined with only one β chain and vice versa.