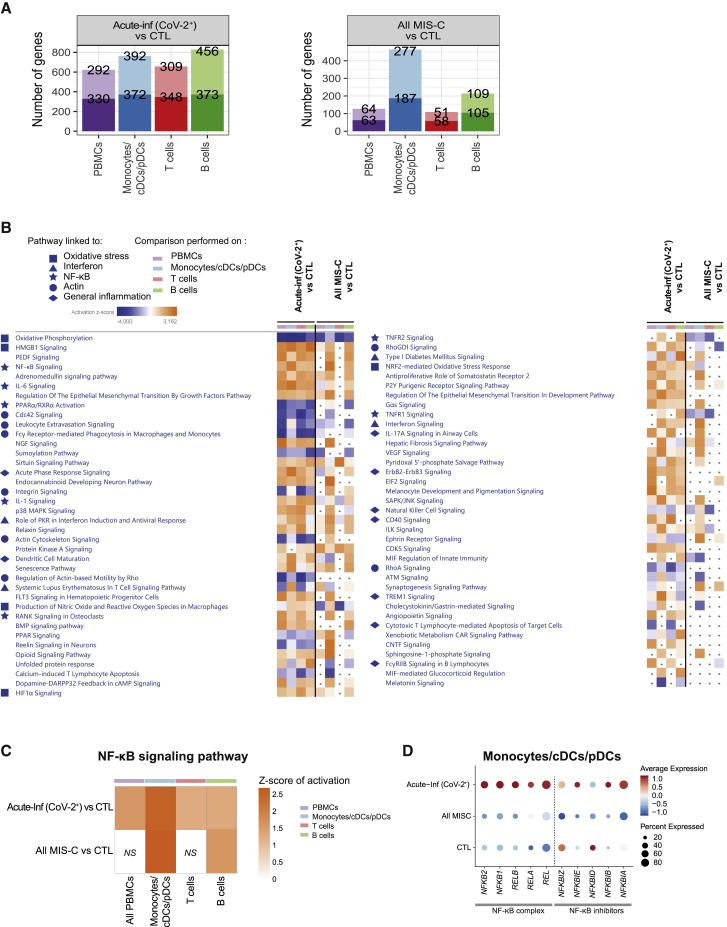
Figure 4.
Genes and pathways differentially regulated in acute infection and postacute hyperinflammation following SARS-CoV-2 infection
(A) Bar charts of the number of up- and downregulated genes in Acute-Inf (CoV-2+) (left panel) and All MIS-C (MIS-C (CoV-2+) and MIS-C_MYO (CoV-2+)) (right panel) compared to CTL in PBMCs, monocyte/cDC/pDC, and T and B cell clusters obtained following scRNA-seq experiments as displayed in Figure 3B. PBMCs represent all clusters. Monocytes/cDCs/pDCs represent clusters 5, 11, 12, 17, 20, 21, and 24. T cells represent clusters 0, 1, 2, 4, 6, 7, 10, 13, 14, 15, 16, 18, and 23. B cells represent clusters 3, 8, 9, 19, and 22. The top value on the light-colored bars represents the upregulated genes, and the bottom value (dark) represents the downregulated genes. Median age for each group: CTL, 15 years; MIS-C (CoV-2+), 3.7 years; MIS-C_MYO (CoV-2+), 8.4 years.
(B) Heatmap of the canonical pathways, enriched in the differentially expressed genes (DEGs) from the comparisons performed in (A) in PBMCs, monocytes/cDCs/pDCs, and T and B cells, obtained by using Ingenuity Pathways Analysis (IPA). Left panel: part 1. Right panel: part 2. Symbols are used in front to represent pathways of the same functional groups. Pathways with an absolute Z score ≤ 2 or adjusted p > 0.05 under all conditions were filtered out. Z score > 2 means that a function is increased significantly (orange) whereas Z score < −2 indicates a significantly decreased function (blue). Gray dots indicate non-significant pathways (p > 0.05).
(C) Heatmap of activation of the NF-κB signaling pathway, as predicted by IPA, in Acute-Inf (CoV-2+) and in All MIS-C (MIS-C (CoV-2+) and MIS-C_MYO (CoV-2+)) compared with controls. The color scale represents the Z score of the prediction. The higher the score, the more activated the NF-κB signaling pathway. NS, non-significant comparison.
(D) Dot plot of the expression in monocytes/cDCs/pDCs of the negative regulators of the NF-κB complex. The color scale shows scaled average expression in all monocytes/cDCs/pDCs, with red and blue being the highest and lowest expression, respectively. Sizes of dots show the percentage of cells that express the gene.