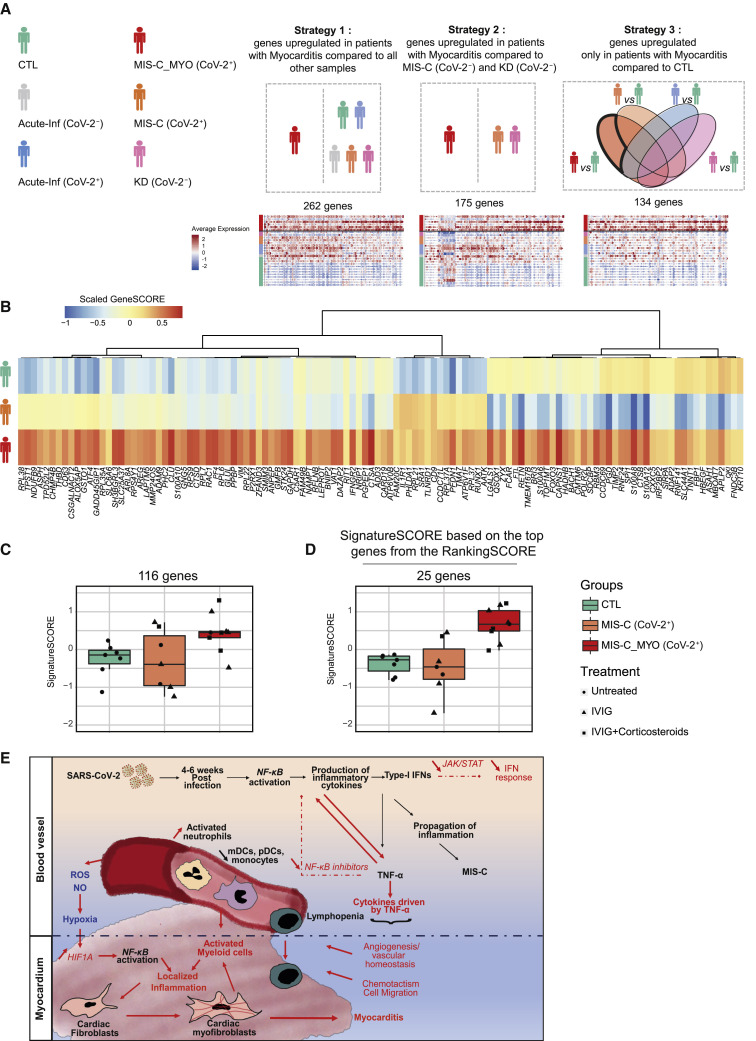
Figure 7.
Molecular signature and proposed mechanism associated with severe myocarditis in children with MIS-C
(A) Schematic of the 3 strategies used to extract 329 markers of the MIS-C_MYO (CoV-2+) group from the monocyte/cDC/pDC clusters using the single-cell dataset. Strategy 1: direct comparison of the monocytes/cDCs/pDCs of the MIS-C_MYO (CoV-2+) group with all other samples. Strategy 2: direct comparison of the monocytes/cDCs/pDCs of MIS-C_MYO (CoV-2+) with other samples with postacute hyperinflammation (MIS-C (CoV-2+) and KD (CoV-2−)). Strategy 3: selection of genes upregulated only in the monocytes/cDCs/pDCs of MIS-C_MYO (CoV-2+) compared with CTL. Below each strategy is the corresponding dot plot obtained from scRNA-seq with the number of upregulated genes. The average expression is represented by the centered scaled expression of each gene. On the left, names of each group with their corresponding color are shown.
(B) Heatmap of expression of the 116 of 329 genes with a higher expression in MIS-C_MYO (CoV-2+) than in other groups in the bulk dataset generated from PBMCs (7 CTL, 7 MIS-C (CoV-2+), 9 MIS-C_MYO (CoV-2+), and 9 KD (CoV-2−) donors). The color scale indicates the scaled GeneSCORE (mean Z score of the gene in all samples of a group), with red and blue representing the highest and lowest expression, respectively. Hierarchical clustering of the genes was computed with a Pearson’s correlation as a distance.
(C) Boxplot of the expression of the 116 genes validated in (C), calculated as a SignatureSCORE, which represents the mean Z score in each sample of the 116 genes selected in (B) in the bulk RNA-seq dataset (Figure S7A).
(D) Boxplot of the SignatureSCORE of the top 25 genes, as ranked in Figure S7B, in the bulk RNA-seq dataset.
(E) Graphical representation based on cytokines,cellular, and transcriptomics analyses (above the black dotted line), combined with known literature (below the black dotted line), illustrating a putative model explaining the occurrence of myocarditis among children in the MIS-C (CoV-2) group. Black represents genes and functions modulated in the MIS-C (CoV-2+) and MIS-C_MYO (CoV-2+) groups compared with CTL, whereas red highlights genes and pathways differentially modulated in the MIS-C (CoV-2+) and MIS-C_MYO (CoV-2+) groups, respectively. See also Figure S7 and Data S3.
(C and D) Each mark represents a sample. Dots represent untreated samples, triangles represent IVIG-treated samples, and squares represent IVIG- and steroid-treated individuals. Boxes range from the 25th to the 75th percentiles. The upper and lower whiskers extend from the box to the largest and smallest values, respectively. Any sample with a value at most ×1.5 the inter-quartile range of the hinge is considered an outlier and plotted individually.