Figure 3. Discovery of 2.6 Mb AR inversion in CTC DNA from Patient 7.
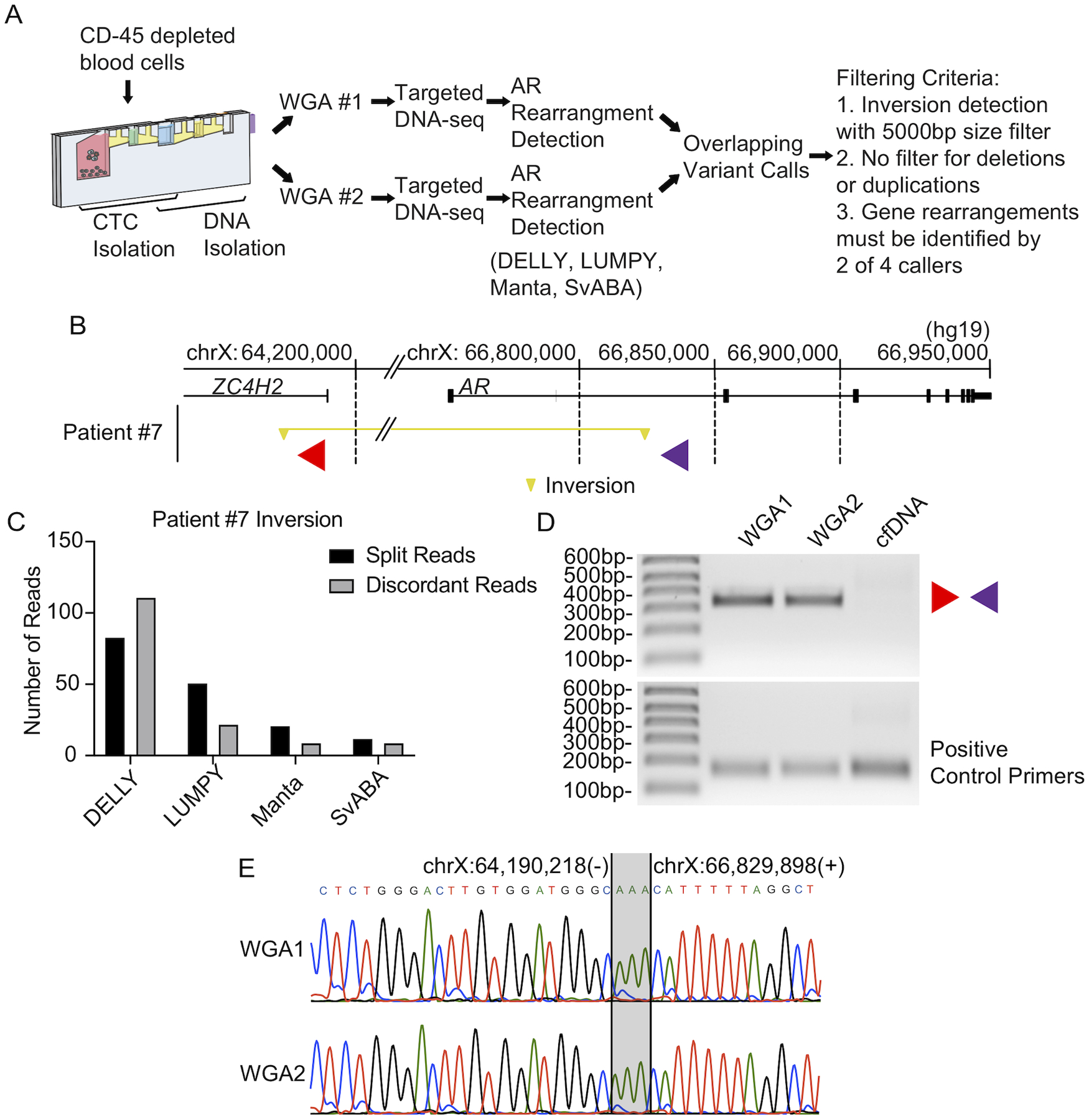
(A) AR gene rearrangement detection pipeline used for circulating tumor cells. (B) Locations of AR gene rearrangement breakpoints caused by a 2.6 Mb inversion in CTC DNA from Patient #7 are shown by yellow triangles. The inverted segment is indicated by a yellow horizontal line. Locations and direction of PCR primers used for validation are denoted with red and purple triangles. Genome coordinates are genome build hg19. (C) Split read counts, discordant read counts, and SV callers supporting the inversion called in (B). Note: LUMPY and Manta made this call as a BND. (D) PCR validating the inversion illustrated in (B) in two parallel WGA reactions of CTC DNA (WGA1 and WGA2), as well as cfDNA from Patient #7. Primers used for PCR are illustrated in (B). (E) Sanger sequencing analysis of the AR inversion PCR products from (D). Microhomology at this breakpoint is indicated with gray shading.
