Fig. 7. DNA damage contributes to CLIP-cohibin disassembly and Cdc48-dependent nucleolar rDNA release.
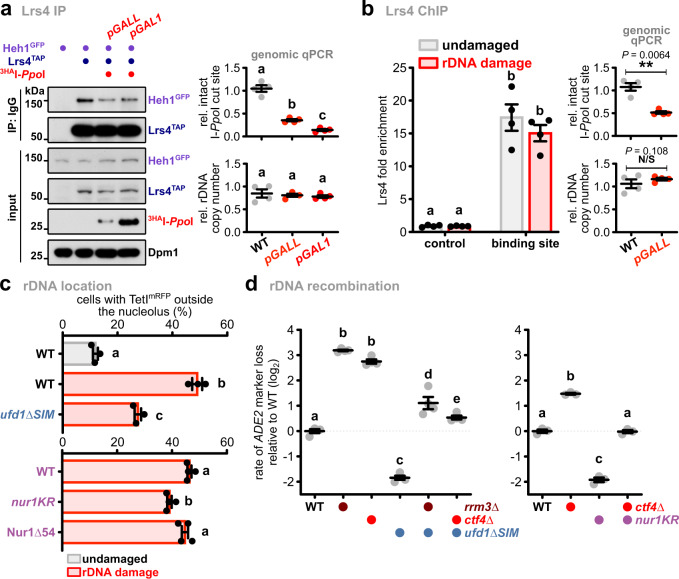
a Co-immunoprecipitation of Heh1GFP with Lrs4TAP in cells with or without rDNA damage generated by 3HAI-PpoI expression. Dpm1 served as loading control. Quantification of double-strand break (DSB) induction (top right) and rDNA copy number (bottom right) relative to undamaged cells is shown (n = 4 independent biological experiments). b Lrs4TAP binding to rDNA repeats in cells with or without rDNA damage, quantified by ChIP-qPCR (n = 4 independent biological experiments). The ChIP values are shown as Lrs4 fold enrichment over the average of three rDNA positions, after normalization to input. Quantification of double-strand break (DSB) induction (top right) and rDNA copy number (bottom right) relative to undamaged cells is shown (n = 4 independent biological experiments), and the statistical analysis was performed using two-tailed Student’s t-test. c Percentage of WT and ufd1∆SIM cells (top; n = 3) or expressing Nur1 full-length (WT), the SUMOylation deficient mutant (nur1KR) or lacking its last 54 residues (Nur1∆54) (bottom; n = 4), with rDNA repeats localized outside the nucleolus before and after DSB induction, quantified as in Fig. 2. d Quantification of rDNA recombination rates in the indicated strains (left, n = 4; right, n = 3). The loss of an ADE2 marker inserted into rDNA is calculated as in Fig. 3, and shown in log2 scale relative to WT. c, d Data are mean ± SEM of n biologically independent experiments. Statistical analysis was performed using ANOVA, and letters denote significant differences with a Tukey’s post hoc test at P ≤ 0.05. Source data are provided as a Source Data file.