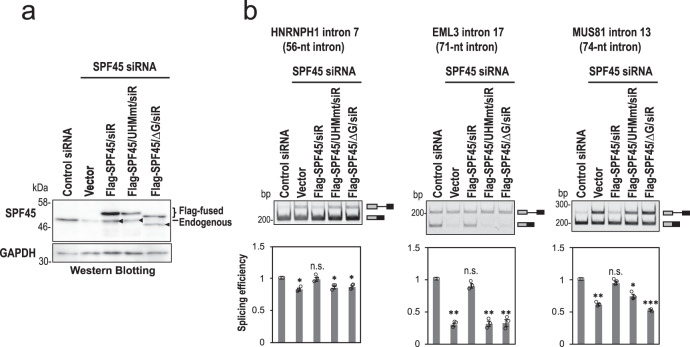
Fig. 8. The expression of siRNA-resistant SPF45 proteins rescue splicing of short introns in SPF45-depleted cells.
a The expressed Flag-fused siRNA-resistant (siR) proteins and endogenous SPF45 in HeLa cells were checked by Western blotting. Displayed blots are representative of three independent experiments. Arrowheads indicate degraded siRNA-resistant (siR) proteins, but not endogenous SPF45 (see ‘vector’ lane for the depletion efficiency of endogenous SPF45). b After the co-transfection of the indicated siRNA-resistant plasmids and three mini-gene plasmids, splicing efficiencies of the indicated three mini genes were analyzed by RT-PCR. Means ± SEM are given for three independent experiments and two-tailed Student t-test values were calculated (HNRNPH1 intron: p = 0.0175 for Control vs Vector, p = 0.8177 for Control vs Flag-SPF45/siR, p = 0.0270 for Control vs Flag-SPF45/UHMmt/siR, p = 0.0250 for Control vs Flag-SPF45/ΔG/siR; EML3 intron: p = 0.0012 for Control vs Vector, p = 0.1085 for Control vs Flag-SPF45/siR, p = 0.0035 for Control vs Flag-SPF45/UHMmt/siR, p = 0.0057 for Control vs Flag-SPF45/ΔG/siR; MUS81 intron: p = 0.0020 for Control vs Vector, p = 0.1693 for Control vs Flag-SPF45/siR, p = 0.0177 for Control vs Flag-SPF45/UHMmt/siR, p = 0.0004 for Control vs Flag-SPF45/ΔG/siR). *P < 0.05, **P < 0.01, ***P < 0.001, n.s.P > 0.05. Source data of all the above panels are provided as a Source Data file.