Abstract
Neurodegenerative dementias are a group of diseases with highly heterogeneous pathology and complicated etiology. There exist potential genetic component overlaps between different neurodegenerative dementias. Here, 1795 patients with neurodegenerative dementias from South China were enrolled, including 1592 with Alzheimer’s disease (AD), 110 with frontotemporal dementia (FTD), and 93 with dementia with Lewy bodies (DLB). Genes targeted sequencing analysis were performed. According to the American College of Medical Genetics (ACMG) guidelines, 39 pathogenic/likely pathogenic (P/LP) variants were identified in 47 unrelated patients in 14 different genes, including PSEN1, PSEN2, APP, MAPT, GRN, CHCHD10, TBK1, VCP, HTRA1, OPTN, SQSTM1, SIGMAR1, and abnormal repeat expansions in C9orf72 and HTT. Overall, 33.3% (13/39) of the variants were novel, the identified P/LP variants were seen in 2.2% (35/1592) and 10.9% (12/110) of AD and FTD cases, respectively. The overall molecular diagnostic rate was 2.6%. Among them, PSEN1 was the most frequently mutated gene (46.8%, 22/47), followed by PSEN2 and APP. Additionally, the age at onset of patients with P/LP variants (51.4 years), ranging from 30 to 83 years, was ~10 years earlier than those without P/LP variants (p < 0.05). This study sheds insight into the genetic spectrum and clinical manifestations of neurodegenerative dementias in South China, further expands the existing repertoire of P/LP variants involved in known dementia-associated genes. It provides a new perspective for basic research on genetic pathogenesis and novel guiding for clinical practice of neurodegenerative dementia.
Subject terms: Disease genetics, Alzheimer's disease, Disease genetics
Introduction
Neurodegenerative dementias are a group of clinically heterogeneous diseases with frequently overlapping symptoms, such as multi-cognitive impairments, behavioral changes, and movement deficits1. Alzheimer’s disease (AD) is the most common dementia worldwide, accounting for 60–80% of all dementia cases2. Frontotemporal dementia (FTD) is the second most common cause of neurodegenerative dementia after AD in patients younger than 65 years, responsible for 10.2% of cases3, and dementia with Lewy bodies (DLB) has been reported as being the second most common dementia subtype in older people following AD, accounting for 7.5% of all dementia cases4. However, the etiology of neurodegenerative dementias is still obscure, which is thought to be caused by a combination of ageing, environmental, and genetic factors.
Recently, substantial progress has been made regarding the molecular genetics of neurodegenerative dementias. PSEN1, PSEN2, and APP are recognized as three causative genes for familial AD (FAD), which explains the genetic background of 5–10% of early onset AD (EOAD, younger than 65 years). The estimated mutation frequencies of PSEN1, APP, and PSEN2 in EOAD, are 80%, 15%, and 5%, respectively5. Likewise, FTD is a genetically and pathologically heterogeneous disorder with a higher incidence of familial cases than AD. Genetic etiology has been revealed in ~30–50% of FTD patients with a positive family history6,7. At present, more than 10 genes are related to FTD, and MAPT, GRN, and C9orf72 are the most common, accounting for ~60% of all cases of inherited FTD3. In contrast, the genetic architecture of DLB remains largely elusive8. To date, only three genes have been confirmed to be related to DLB, including APOE, GBA, and SNCA. However, growing evidence supports that DLB has a strong and unique genetic component9.
Interestingly, previous studies have suggested a potential genetic overlap between AD, FTD, and DLB. Notably, PSEN1, the most common etiology of EOAD, has also been found in patients with FTD and DLB10–13. Similarly, mutations in MAPT, GRN, and C9orf72 have also been detected at lower frequencies in AD and DLB patients14–16. Homozygosity for APOE4, the strongest genetic risk factor for AD, has also been reported in several studies to increase the risk of FTD and DLB17,18. In addition, mutations in SNCA have been shown to result in a wide phenotypic spectrum of DLB, Parkinson’s disease (PD), multiple system atrophy (MSA), and FTD19–21.
In this study, we comprehensively analyzed the mutational spectrum of known dementia-associated genes from patients with neurodegenerative dementias in the South Chinese population using integrated targeted gene sequencing analysis. First, we systematically identified pathogenic and likely pathogenic (P/LP) variants of known dementia-associated genes, including known and novel variants, summarized and compared the mutation frequency among patients with different clinical diagnosis. Second, we generalized the clinical manifestation of neurodegenerative dementia patients carried P/LP variants in this study, including PSEN1, PSEN2, APP, MAPT, GRN, C9orf72, CHCHD10, HTRA1, TBK1, OPTN, SQSTM1, VCP, SIGMAR1, and HTT, attempting to summarize the relationship between gene mutations and clinical phenotypes. Then, we compared the age at onset (AAO) of patients with and without P/LP variants and patients carried different genes separately, to depict the AAO spectrum for these dementia-associated genes in our population. Finally, we analyzed APOE genotypes (non-carriers or carriers of APOE4) in AD cohort and conclude the difference between APOE genotypes and different AD subgroups. Our studies provide a new perspective for further basic research of neurodegenerative dementia, especially genetic-associated pathogenesis and facilitated the clinical prediction, diagnosis, and genetic counseling.
Results
Demographics and analysis of genes targeted sequence
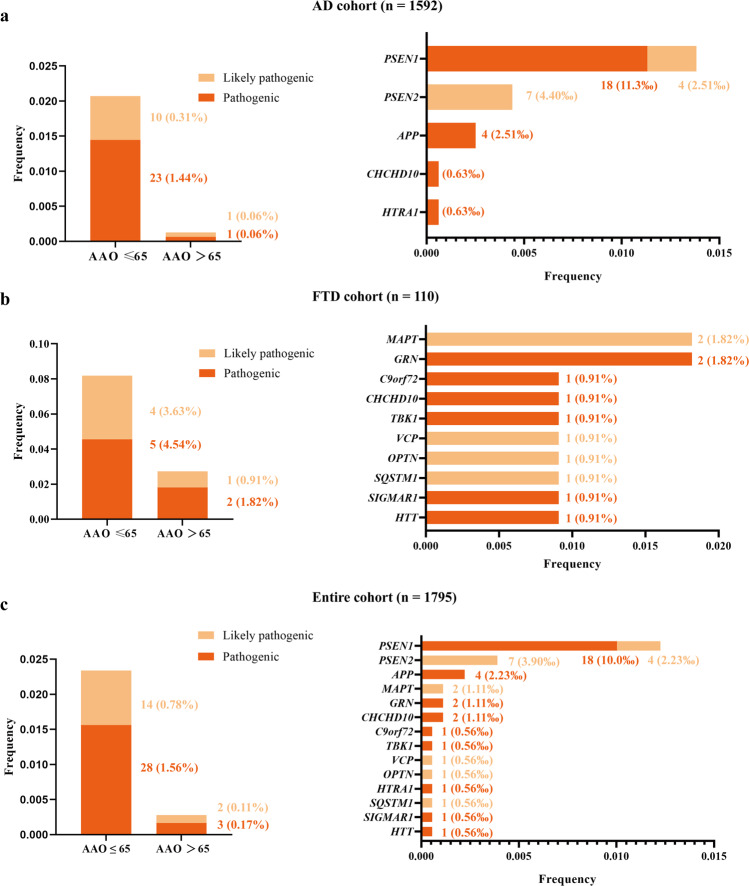
In this study, 1592 AD patients, 110 FTD patients, and 93 DLB patients were included. Demographic and clinical characteristics are shown in Table 1. A total of 39 P/LP variants from 14 genes are identified in 47 unrelated patients by a dementia-related gene panel, which contained 36 genes associated with cognitive impairment phenotype (Supplementary Table 1 and Supplementary 2). Among them, 33.3% of variants (13/39) were novel, including PSEN1 (c.451G>A, c679A>c, c.A1139>G, and c.1369 A>G), PSEN2 (c.T716C and c.1180delG), GRN (c.20G>A), CHCHD10 (c.121C>T and c.283C>T), OPTN (c.1402_1407del), SQSTM1 (c.558_559insC), SIGMAR1 (c.26G>A), and TBK1 (c.973dup). All identified P/LP variants were responsible for 2.2% of AD (35/1592) and 10.9% of FTD (12/110), which led to an overall molecular diagnostic yield of 2.6% (Fig. 1), however, in this study, no P/LP variants were identified in DLB patients. 70.2% (33/47) of patients had a positive family history and 46.8% (22/47) of patients with P/LP variants had at least one APOE4.
Table 1.
Summary of clinical features of the cognitive impairment disease patients in this study.
| Clinical features | AD (n = 1592) | FTD (n = 110) | DLB (n = 93) | Total (n = 1795) |
|---|---|---|---|---|
| Age at onset, years | 64.7 ± 10.8 | 59.6 ± 11.5 | 65.2 ± 9.9 | 64.5 ± 10.9 |
| Gender (M, %) | 645, 40.4% | 56, 50.9% | 57, 61.3% | 758, 42.1% |
| Age at diagnosis, years | 67.7 ± 10.9 | 62.4 ± 11.8 | 68.0 ± 9.6 | 67.4 ± 11.0 |
| Family history (+, %) | 467, 29.3% | 35, 31.8% | 16, 17.2% | 518, 28.8% |
| Disease duration, years | 3.0 ± 2.4 | 2.8 ± 2.3 | 2.9 ± 2.8 | 3.0 ± 2.4 |
| Education attainment, years | 8.5 ± 4.1 | 9.0 ± 4.0 | 7.7 ± 4.0 | 8.5 ± 4.1 |
| MMSE | 13.8 ± 8.5 | 15.3 ± 9.2 | 15.7 ± 7.7 | 14.0 ± 8.5 |
| APOE genotypes (n, %) | ||||
| APOE4 +/+ | 145, 9.1% | 5, 4.5% | 7, 7.5% | 157, 8.7% |
| APOE4 +/− | 561, 35.2% | 36, 32.7% | 29, 31.2% | 626, 34.9% |
| APOE4 −/− | 886, 55.7% | 69, 62.8% | 57, 61.3% | 1012, 56.4% |
The age at onset, the age at diagnosis, disease duration, educational attainment, MMSE scores are all shown as mean ± standard deviation. Gender, family history, APOE genotypes are all shown as numbers and proportions (%).
AD Alzheimer’s disease, FTD frontotemporal dementia, DLB dementia with Lewy bodies, MMSE Mini-Mental State Examination.
Fig. 1. Mutational frequencies of known cognitive impairment disease-associated genes in AD, FTD, and entire cohorts respectively.
Mutational frequencies of all (left) and each (right) known cognitive impairment-associated genes in the different dementia cohorts. a AD cohort. b FTD cohort. c entire cohort. Variants that were classified as pathogenic or likely pathogenic according to the standards and guidelines of the ACMG. ‘Pathogenic’ means that the patients had pathogenic variants in known cognitive impairment disease-associated genes, and ‘likely pathogenic’ means that the patients had likely pathogenic variants in known cognitive impairment disease-associated genes. ACMG American College of Medical Genetics, AD Alzheimer’s disease, FTD frontotemporal dementia.
Mutational spectrum of the AD cohort
Overall, 27 different P/LP variants were identified in 35 unrelated AD patients from five genes, including PSEN1, PSEN2, APP, CHCHD10, and HTRA1 (Table 2). 88.6% (31/35) carrier were FAD probands, 11.4% (4/35) were sporadic AD (SAD) cases.
Table 2.
Clinical characteristics and variants information of patients with pathogenic or likely pathogenic mutations.
| Genes | Base change | Protein change | No. of cases | Reported and references | Gender | AAO (ys) | APOE genotype | Family history | Clinical phenotypes | Clinical diagnosis | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Memory decline | Language impairment | Mental and behavior change | Sensory and movement disorders | ||||||||||
| PSEN1 | c.250A>G | p.M84V | 1 | Y63 | M | 53 | 3/4 | + | + | + | + | + | AD |
| c.415A>G | p.M139V | 1 | Y64 | M | 53 | 2/3 | + | + | + | + | − | AD | |
| c.415A>T | p.M139L | 1 | Y65 | F | 38 | 4/4 | + | + | − | − | + | AD | |
| c.424G>A | p.V142I | 2 | Y66 | M/M | 54/52 | 3/3 | +/+ | +/+ | +/− | −/+ | −/− | AD | |
| c.436A>G | p.M146V | 1 | Y64 | M | 42 | 3/4 | + | + | + | + | − | AD | |
| c.451G>A | p.V151M | 1 | N | M | 49 | 3/4 | + | + | + | + | − | AD | |
| c.519G>T | p.L173F | 1 | Y67 | F | 37 | 3/4 | + | + | − | + | − | AD | |
| c.604A>T | p.I202F | 1 | Y68 | F | 46 | 3/4 | + | + | + | − | − | AD | |
| c.617G>A | p.G206D | 1 | Y69 | M | 38 | 3/3 | + | + | − | + | − | AD | |
| c.677T>G | p.L226R | 1 | Y70 | M | 44 | 3/4 | + | + | − | + | − | AD | |
| c.679A>C | p.I227L | 1 | N | M | 44 | 3/4 | − | + | − | − | − | AD | |
| c.697A>G | p.M233V | 1 | Y71 | M | 30 | 3/3 | + | + | − | − | + | AD | |
| c.791C>T | p. P264L | 2 | Y72 | M/F | 51/52 | 3/4 | +/− | +/+ | +/+ | +/− | +/− | AD | |
| c.806G>A | p.R269H | 2 | Y73 | M/F | 45/60 | 3/4 | +/+ | +/+ | +/− | −/− | −/− | AD | |
| c.845T>G | p.L282R | 1 | Y74 | F | 52 | 3/3 | + | + | + | − | + | AD | |
| c.854C>T | p.A285V | 1 | Y75 | F | 46 | 3/3 | + | + | + | − | − | AD | |
| c.1139A>G | p.K380R | 1 | N | F | 49 | 3/4 | + | + | − | − | − | AD | |
| c.1174C>G | p.L392V | 1 | Y76 | F | 46 | 3/3 | + | + | − | + | − | AD | |
| c.1369A>G | p.M457V | 1 | N | M | 66 | 3/4 | + | + | − | + | − | AD | |
| PSEN2 | c.715A>G | p.M239V | 4 | Y77 | M/M/F/F | 47/60/53/45 |
50% 3/3 50% 3/4 |
+/+/+/+ | +/+/+/+ | −/−/+/− | −/+/+/+ | −/−/−/+ | AD |
| c.716T>C | p.M239T | 1 | N | F | 50 | 3/4 | − | + | − | − | − | AD | |
| c.717G>A | p.M239I | 1 | Y32 | F | 50 | 3/4 | + | + | − | + | − | AD | |
| c.1180delG | p.A394Pfs*8 | 1 | N | F | 68 | 3/3 | + | + | − | + | − | AD | |
| APP | c.2143G>A | p.V715M | 1 | Y78 | F | 51 | 3/3 | + | + | − | + | − | AD |
| c.2149G>A | p.V717I | 3 | Y79 | F/F/M | 47/49/42 | 66.7% 3/3; 33.3% 3/4 | +/+/+ | +/+/+ | +/+/+ | −/+/+ | −/−/− | AD | |
| MAPT | c.1788T>G | p.N596K | 1 | Y80 | F | 42 | 3/3 | − | + | − | − | + | FTD |
| c.1907C>T | p.P636L | 1 | Y81 | M | 58 | 3/3 | − | + | + | + | − | FTD | |
| GRN | c.20G>A | p.W7* | 1 | N | F | 73 | 3/4 | + | + | − | + | − | FTD |
| c.328C>T | p.R110* | 1 | Y82 | F | 61 | 3/3 | − | + | + | + | − | FTD | |
| CHCHD10 | c.283C>T | p.Q95* | 1 | N | F | 52 | 3/4 | + | + | + | + | + | AD |
| c.121C>T | p.Q41* | 1 | N | F | 56 | 3/4 | − | + | − | + | − | FTD | |
| HTRA1 | c.589C>T | p.R197* | 1 | Y83 | F | 49 | 3/3 | − | + | − | + | − | AD |
| OPTN | c.1402_1407del | p.468_469del | 1 | N | F | 63 | 3/3 | − | + | + | − | − | FTD |
| SQSTM1 | c.558_559insC | p.V287Rfs*21 | 1 | N | F | 71 | 3/3 | − | + | + | − | − | FTD |
| VCP | c.475C>T | p.R159C | 1 | Y84 | M | 51 | 3/3 | − | + | − | + | − | FTD |
| SIGMAR1 | c.26G>A | p.W9* | 1 | N | F | 74 | 3/3 | − | + | − | + | − | FTD |
| TBK1 | c.973dup | p.Y325Lfs*4 | 1 | N | M | 61 | 3/3 | − | + | + | + | − | FTD |
| C9orf72 | Hexanucleotide expansion | − | 1 | Y | F | 57 | 3/4 | + | + | + | − | + | FTD |
| HTT | CAG repeat expansion | – | 1 | Y | F | 43 | 3/3 | + | + | + | − | + | HD |
Y mutations have been reported previously, N mutations have not been reported previously.
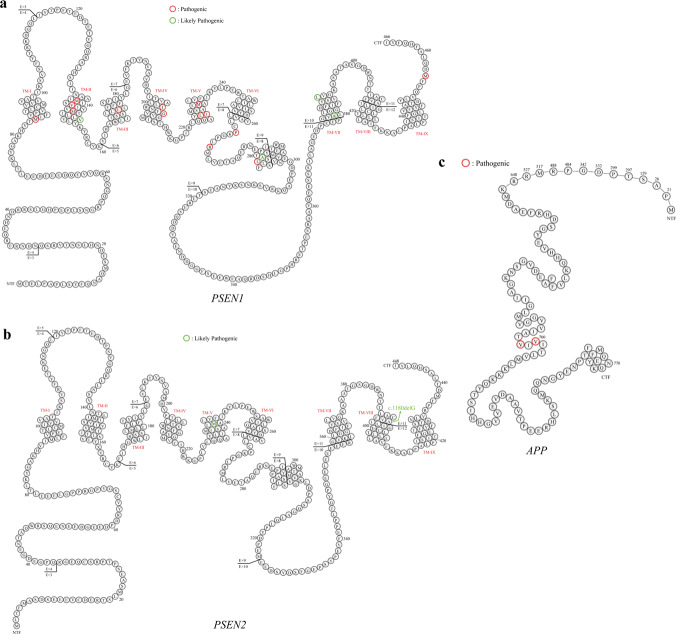
In this study, PSEN1 was the most frequently mutated gene, 19 P/LP missense mutations were identified in 22 patients, among which four were novel identified in our study, including c.451G>A, p.V151M; c.679A>C, p.I227L; c.1139A>G, p.K380R; and c.1369A>G, p.M457V. Seven patients carried PSEN2 P/LP variants, including six missense mutations at the same amino acid residue (M239) and one frameshift mutation. Two were novel, including c.T716C, p.239M>T and c.1180delG, p.A394Pfs*8. All patients with the variants had a positive family history except for one who carried PSEN2 p.239M>T. Meanwhile, two APP missense mutations were identified in four FAD probands, including c.2143G>A, p.V715M, and c.2149G>A, p.V717I (Table 2). The distribution of PSEN1/PSEN2/APP P/LP variants are shown in Fig. 2. Interestingly, all identified P/LP variants of PSEN1/PSEN2/APP were located in hydrophobic regions or in the endoproteolytic cleavage regions.
Fig. 2. Distribution of amino acid substitutions in the PSEN1, PSEN2, and APP proteins.
Red circles: pathogenic mutations identified. Green circles: likely pathogenic mutations identified. A Ala, C Cys, D Asp, E Glu, F Phe, G Gly, H His, I Ile, K Lys, L Leu, M Met, N Asn, P Pro, Q Gln, R Arg, S Ser, T Thr, V Val, W Trp, Y Tyr, APP amyloid precursor protein, PSEN presenilin. a The distribution of PSEN1 P/LP variants. b The distribution of PSEN2 P/LP variants. c The distribution of APP P/LP variatns.
In patients with PSEN1/PSEN2/APP variants, 93.9% (31/33) were defined as early-onset AD (EOAD) (AAO < 65 years)), 45.5% were APOE4-negative, 48.5% had one APOE4, and 6.0% had two copies of APOE4.
About clinical phenotypes, all PSEN1/PSEN2/APP variants carriers initially presented with memory decline. Then, language impairment and behavior change were common symptoms in these variants, 52.63% (10/19) PSEN1, 25% (1/4) PSEN2, and 50% (1/2) APP P/LP variants showed language impairment, respectively, such as naming difficulty, repetitive speech, fluency disorder, and speech reduction, while the frequency of mental and behavior change of PSEN1, PSEN2, APP variants were 57.6% (11/19), 75% (3/4), and 100% (2/2), respectively. Meanwhile, 26.3% (5/19) PSEN1, 25% (1/4) PSEN2 P/LP variants presented sensory and movement disorders, such as hallucination, delusion, weakness, involuntary movement, and abnormal gait. Interestingly, the clinical manifestation of patients with PSEN2 mutations at amino acid residue M239 showed high heterogeneity, including memory decline, language impairment, mental and behavior change, and sensory and movement disorders.
Additionally, we also found two female AD patients who carried a nonsense mutation in CHCHD10 (c.283C>T, p.Q95*) and HTRA1 (c.589C>T, p.R197*), respectively. The patient who carried CHCHD10 p.Q95* showed memory decline at 52 years and gradually developed language dysfunction, behavioral changes, bradykinesia, and depression. Brain MRI showed bilateral atrophy of temporal parietal lobe and hippocampus, and cerebrospinal fluid (CSF) examination showed the level of Aβ42 and Aβ42/Aβ40 ratio decreased, while the phospho-tau (p-tau) and total tau (t-tau) increased. In addition, the Pittsburgh compound B (PiB)-PET showed diffuse amyloid deposition in the whole brain cortex. The patient who carried HTRA1, p.R197*, mainly presented typical forgetfulness of recent events and daily living ability declined at 49 years. Brain MRI showed multiple spot-like hyperintensities in the deep bilateral frontotemporal lobes and paraventricular region, while no microbleeds on susceptibility-weighted images sequence. The level of Aβ42 in CSF decreased, and p-tau increased which supported the diagnosis of AD.
Mutational spectrum of the FTD cohort
A total of 12 P/LP variants in 10 genes were identified in the FTD cohort, including MAPT, GRN, C9orf72, CHCHD10, TBK1, OPTN, SQSTM1, VCP, SIGMAR1, and HTT, summarized in Table 2. Six were novel variants, including three nonsense mutations (GRN: c. 20G>A, p.W7*; CHCHD10: c. 121C>T, p.Q41*; and SIGMAR1: c.26G>A, p.W9*), two frameshift mutations (TBK1: c.973dup, p.Y325Lfs*4 and SQSTM1: c.558_559insC, p.V287Rfs*21), and one deletion mutation (OPTN: c.1402_1407del, p.468_469del). Interestingly, only two P/LP variants carriers had a positive family history. The mean ± SD AAO of P/LP variants carriers was 59.2 ± 10.5 years, which was significantly older than P/LP variants carriers in AD cohort (48.8 ± 7.7, p = 0.001). Meanwhile, only three patients carried APOE4 (25%, 3/12), which tended to be lower than P/LP variant carriers in the AD cohort (54.3%, p = 0.077).
As for clinical characteristics, all patients carried P/LP variants in FTD cohort showed memory decline, 58.33% (7/12) patients had language impairment, mental, and behavior changes, and 25% (4/12) P/LP variants carriers accompanied by sensory and movement disorders. Interestingly, one showed personality changes and language impairment, as well as abnormal emotional responses at baseline. In the fifth year of onset, she suffered from memory decline. Brain MRI showed bilateral frontal lobe atrophy, and bvFTD was initially considered. However, molecular testing revealed that she carried heterozygous CAG expanded repeats in HTT, which supported the diagnosis of Huntington’s disease (HD).
Spectrum of age at onset
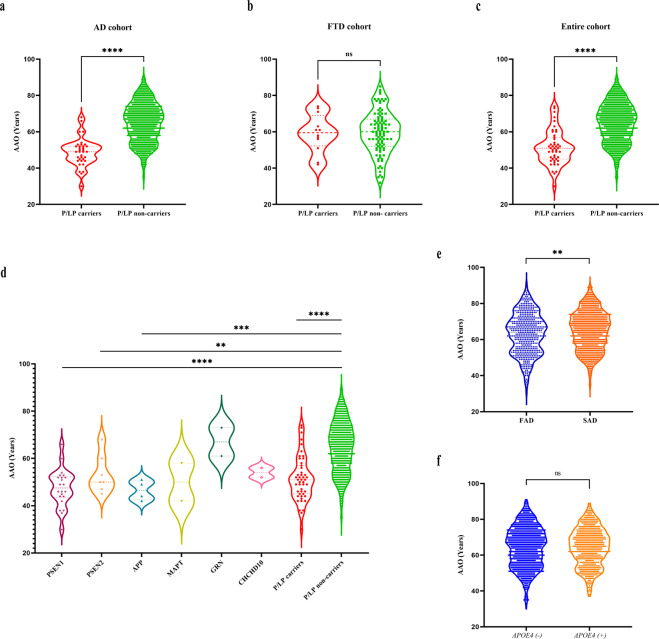
Moreover, the mean AAO were significantly younger in patients with P/LP variants in the AD cohort and entire cohort, while no difference was found in the FTD cohort (Fig. 3a–c). Specifically, the mean AAO of patients with P/LP variants in the entire cohort was 51.4 ± 9.5 years, ~10 years younger than the mean AAO of those without P/LP variants (64.8 ± 10.7 years) (p < 0.001), among them, 89.4% were younger than 65 years.
Fig. 3. AAO spectrum of known cognitive impairment disease-associated genes in AD, FTD, and entire cohorts.
Comparison of AAO in all patients with P/LP variants of known cognitive impairment disease-associated genes and patients without P/LP variants in known cognitive impairment disease-associated genes. a AD cohort. b FTD cohort. c Entire cohort. The dashed red line refers to the mean AAO of patients with P/LP variants in the corresponding cohorts, whereas the dashed line refers to the mean AAO of patients without P/LP variants in known cognitive impairment disease-associated genes in the corresponding cohort. d Spectrum of AAO in patients with P/LP variants of each cognitive impairment disease-associated gene (only genes carried by two or more patients were included), in patients with and without P/LP variants of known cognitive impairment disease-associated genes. e Spectrum of AAO in FAD and SAD patients. f Spectrum of AAO in patients with and without APOE4. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns no significance. AAO age at onset, AD Alzheimer’s disease, FTD frontotemporal dementia, P/LP pathogenic or likely pathogenic, FAD familial Alzheimer’s disease, SAD sporadic Alzheimer’s disease.
Meanwhile, we analyzed the spectrum of AAO in patients with P/LP variants of different genes (genes with two or more mutations were included). The results showed that the mean AAO of subjects with P/LP variants of PSEN1, PSEN2, and APP (47.5 ± 10.7 years, 53.2 ± 8.1 years, and 46.5 ± 4.2 years, respectively) were significantly lower than those of non-carriers (p < 0.05) (Fig. 3d).
In addition, we performed subgroup analysis on the family history and the status of APOE4 to compare the difference in AAO between the two groups respectively, which showed that the AAO of FAD patients was significantly younger than that of SAD (63.2 ± 11.4 and 64.9 ± 10.6, respectively, p = 0.005), while no significant between APOE4 carriers and non-carriers (p = 0.953) (Fig. 3e, f).
To analyze the confounding factors affecting AAO, we conducted further multiple linear regression analysis with AAO as the dependent variable. After controlling independent variables, including gender, disease duration, educational attainment, APOE genotypes, dementia family history, MMSE scores, mutation status, and clinical diagnosis, the model showed that MMSE scores (B = −0.135, p < 0.001), disease duration (B = −0.421, p = 0.001), and status of mutation carried (B = −13.44, p < 0.001).
Characteristics of APOE genotypes
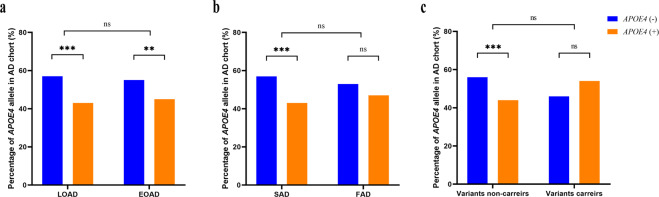
As for the distributions of APOE genotypes, there was no significant difference across AD, FTD, and DLB cohorts (p > 0.0166; Bonferroni corrected). Furthermore, APOE4 as the strongest genetic risk factor for AD, we further compared the distribution difference between EOAD and LOAD patients, FAD and SAD. We found no significant difference in APOE4 frequency (EOAD vs LOAD: p = 0.501; FAD vs SAD: p = 0.153, respectively). Further subgroup analysis in AD cohort showed that the proportion of APOE4-negative patients was higher than that of APOE4-positive patients (p < 0.001, p = 0.007, and p < 0.001, respectively) (Fig. 4a, b). In addition, we found no significant difference in the distribution difference of APOE4 between variants carriers and non-carriers in the AD cohort (p = 0.281). Meanwhile, a higher percentage of APOE4 (+) patients was found in P/LP variants than in patients without P/LP variant in AD cohort (p < 0.001) (Fig. 4c).
Fig. 4. The percentage of APOE4 in AD cohort.
a The percentage of APOE4 in LOAD and EOAD. b The percentage of APOE4 in SAD and FAD. c The percentage of APOE4 in AD patients with and without P/LP variants. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns no significance. LOAD late-onset Alzheimer’s disease, EOAD early-onset Alzheimer’s disease, SAD sporadic Alzheimer’s disease, FAD familial Alzheimer’s disease.
Discussion
In this study, we determined the mutational spectrum of 36 known dementia-associated genes in patients clinically diagnosed with neurodegenerative dementia patients, including AD, FTD, and DLB in a South China population sample using integrated targeted gene sequencing analysis. This is the first report of distributions of gene mutations in patients with neurodegenerative dementias from South China. We observed that the use of an integrated gene analysis could be an effective tool for detecting potential genetic causes in neurodegenerative dementias with high genetic heterogeneity or overlapping phenotypic features, gaining further insight in genetic pathogenesis of neurodegenerative dementia, clinical diagnosis, and genetic counseling.
Mutations in PSEN1 are the most common cause of EOAD, meanwhile, PSEN1 was the most frequently mutated gene in patients with FAD. To date, more than 300 mutations in PSEN1 have been identified to be associated with FAD. In this study, four novel variants were identified, which expanded the mutational spectrum of PSEN1. The AAO of PSEN1 mutation carriers in our study (47.5 years), was older than Ryan et al. reported (43.6 years) in 168 AD patients with PSEN1 mutations22, but younger than Jia et al. reported in a large FAD cohort from China (50.59 years)23. Of interest, in addition to the PSEN1 mutations mentioned above, we found an older female (83 years) carrying a novel mutation (M270L), and the APOE genotype was 3/4 in the DLB cohort. Several algorithms predicted the variant was not disease damaging, whereas the nearby mutations (R269G, R269H, and L271V) have been reported to be associated with FAD24–26. Whether the clinical phenotype of the patient is caused by the novel mutation or the contribution of APOE genotype is unclear; we will perform functional research to further clarify the variant. Meanwhile, regarding clinical phenotypes, PSEN1 mutation carriers often present with atypical cognitive symptoms and additional neurological features22. However, in this study, patients mainly presented with amnesia, language impairment, mental and behavioral changes, and movement disorders, which is one limitation of this study. This might have two explanations. First, some atypical symptoms might not occur at an early stage of the disease, and follow-up is necessary. Second, the mutation locations may lead to distinguishing phenotypes; for example, atypical cognitive presentations and pyramidal signs were seen more frequently in association with PSEN1 mutations involving exon 822, suggesting that multiple factors could contribute to the phenotypic heterogeneity of PSEN1-related AD.
In contrast to PSEN1, only 18 pathogenic mutations within PSEN2 have been reported, most of which occurred in European and African populations. In this study, seven P/LP variants were identified, including six FAD cases. Previous studies showed that the AAO of PSEN2-associated cases vary widely, from 45 to 88 years; that is more than 10 years later than the mean AAO for PSEN1-related cases27–29, which was consistent with our results. Interestingly, six patients with a substitution at PSEN2 amino acid residue 239 were identified, including M239V, M239I, and M239T. Among them, M239V has been reported in European populations to elevate Aβ42 levels and Aβ42/Aβ40, and to exhibit a partial loss of function with respect to the C-terminal fragment-γ as well as a substantial decrease in Aβ40 levels30–32, but it has been absent from Asian cohorts so far. Our findings suggest that this residue may be a common causative variant in the South Chinese population. In addition, the clinical phenotypes of carriers of the M239V mutation varied widely. Our findings, together with previous reports, further suggest that phenotypic heterogeneity exists even at the same codon site because of different amino acid transversions30,32,33.
In accordance with other populations, the common mutation site of APP were residues 715 and 717. Taken together with our previous reports, our team have found four families carrying mutations at this site34. Amino acid residues 715 and 717 are located near the γ-secretase cleavage site, and mutation at this site may increase the hydrophobicity of the APP TM domain to anchor the protein within the membrane and elevate the Aβ42/Aβ40 ratio35,36. Interestingly, patients with mutations at this site often have non-memory symptoms, which can be misdiagnosed as FTD, because the behavioral problems occur earlier than the memory deficits. Totally, in all PSEN1/PSEN2/APP P/LP variants in LOAD, PSEN1 M457V, and PSEN2 A394Pfs*8 were novel variants, further functional validation was necessary and warranted.
In addition, we identified CHCHD10 and HTRA1 mutations in the AD cohort. CHCHD10 has been identified to be associated with a large spectrum of diseases, including FTD, ALS, AD, cerebellar ataxia, mitochondrial myopathy, late-onset spinal motor neuronopathy, and Charcot-Marie-Tooth disease type 237–40. Previously, we have reported a late-onset AD patient with the CHCHD10 mutation41. A homozygous HTRA1 mutation was known to be causative for CARASIL42, while evidence was also showed that heterozygous HTRA1 mutation, which might result in an impaired HTRA1 activation cascade or be unable to form stable trimers, is related to autosomal dominant hereditary cerebral small vessel disease with delayed onset43–45. In this study, the female AD patients presented with typical amnesia symptoms at 49 years without any other neurological or extra-neurological symptoms. Meanwhile, the Fazekas score of periventricular white matter hyperintensities was 1, the APOE genotype was 3/3 and the core biomarkers of CSF showed A+T+N-. However, the effect of the heterozygous mutation in the pathophysiologic process of AD remains elusive; further functional studies are still needed. These results further indicate that mutations not only in PSEN1, PSEN2, and APP can cause the AD phenotype, but that variants in other genes might also cause AD-like symptoms. Further follow-up is necessary.
Notably, we identified double mutations in a 52-year-old female (PSEN2 p.M239I and MAPT p.R5H), but her daily living ability remained intact, and the double mutation did not accelerate the cognitive decline, further expanding the phenotype spectrum of the mutation and supporting the phenotypic heterogeneity among subjects carrying the same MAPT mutations. Further in vivo and in vitro studies are needed to determine the effect of MAPT and PSEN2 mutations on the pathology and pathogenesis of AD.
Many different gene are reported to cause FTD, of which MAPT, GRN, and C9orf72 are three most common46–48. Except of three common genes, we also found variants in another seven genes, including CHCHD10, OPTN, SQSTM1, VCP, SIGMAR1, TBK1, and HTT49–52. In addition to genetics, the clinical phenotypes of FTD are also highly heterogeneous. In this study, we did not observe the classic phenotypes of mutations in VCP, such as inclusion body myopathy with Paget’s disease of the bone53,54, we will follow up the patient to see the symptoms evolve. Moreover, the wrong diagnosis of the patient carried heterozygous CAG expanded repeats in HTT, further indicated that the overlap of clinical phenotypes is one of the main reasons for the difficulty in the diagnosis of neurodegenerative diseases. Genetic analysis is an effective method to improve diagnostic certainty.
Additionally, in our study, the proportion of APOE4 positive cases between FAD and SAD, EOAD and LOAD, were not significantly different, which is inconsistent with previous studies reporting that APOE4 exerts its maximal effect in EOAD55,56. Perhaps other genetic or environmental factors may play an important role in the onset and pathogenesis of AD.
This study represents a comprehensive and systematic screening of 36 dementia-associated genes in AD, FTD, and DLB patients from South China, although the current study has some limitations. First, we only focused on known 36 dementia-associated genes, not susceptibility genes, risk loci, or new candidate genes, which may play important roles in neurodegenerative dementia. Second, in this study, we only screened neurodegenerative dementia patients, but no controls were assessed to compare background frequencies of the P/LP variants. Lastly, for those novel variants identified in this study, we did not design functional experiments to further validation.
In conclusion, we have conducted the most systematic survey of the mutational spectrum of neurodegenerative dementia patients in South Chinese population, which further expanded the mutational spectrum of dementia-related genes and have provided evidence that there is some genetic heterogeneity and perhaps overlap between phenotypes. Our results may prove to be beneficial for clinical prediction, diagnosis, and genetic counseling and may generate hypotheses for future basic research on genetic-associated pathogenesis of neurodegenerative dementia.
Methods
Study participants
A total of 1795 patients with neurodegenerative dementias, including 1592 with AD, 110 with FTD, and 93 with DLB, were recruited at the Xiangya Hospital, Central South University, between February 2004 and October 2020. All patients were unrelated probands. The demographic and clinical characteristics are summarized in Table 1. All subjects had been clinically diagnosed with AD, FTD, or DLB according to international guidelines. This study was approved by the Ethics Committee of Xiangya Hospital, Central South University, China. Written informed consent was obtained from each participant or guardian.
Targeted genes sequencing
Genomic DNA was extracted from peripheral blood leukocytes of each participant using the QIAGEN kit according to the manufacturer’s instructions. We designed a dementia-related gene panel containing a total of 36 genes associated with cognitive impairment phenotypes, including PSEN1, PSEN2, APP, APOE, ABCA7, SORL1, TREM2, ADAM10, MAPT, GRN, FUS, TARDBP, VCP, TBK1, CHCHD10, HTRA1, SQSTM1, UBQLN1, CHMP2B, SIGMAR1, OPTN, HNRNPA1, HNRNPA2B1, PRKAR1B, TMEM106B, UBQLN2, NOTCH3, TREX1, GLA, COL4A1, CSF1R, GBA, SNCA, SNCB, LRRK2, and PRNP. Briefly, gDNA was fragmented and a paired-end library was constructed using Covaris LE220 (Massachusetts, USA), followed by pre-capture PCR amplification. After PCR amplification, the DNA fragments were captured by the targeted panel, followed by sequencing using the Illumina NovaSeq 6000 platform. The reads were mapped to the human genome reference (hg19) using the Burrows Wheeler Aligner software (http://bio-bwa.sourceforge.net)57, and duplicate sequence reads were removed using Picard (http://broadinstitute.github.io/picard/). Variant calling was performed using the Genome Analysis Toolkit (https://software.broadinstitute.org/gatk/)58. The variants were annotated using ANNOVAR (https://hpc.nih.gov/apps/ANNOVAR.html)59 and named according to the guidelines of the Human Genome Variation Society (http://www.hgvs.org/)60. Pathogenic or likely pathogenic (P/LP) variants were assessed according to the guidelines issued by the American College of Medical Genetics (ACMG).
Moreover, the (GGGGCC)n repeats in C9orf72 and (CAG)n repeats in HTT were performed in all individuals using previously reported repeat-primed polymerase chain reaction and capillary electrophoresis61,62.
Sanger sequencing
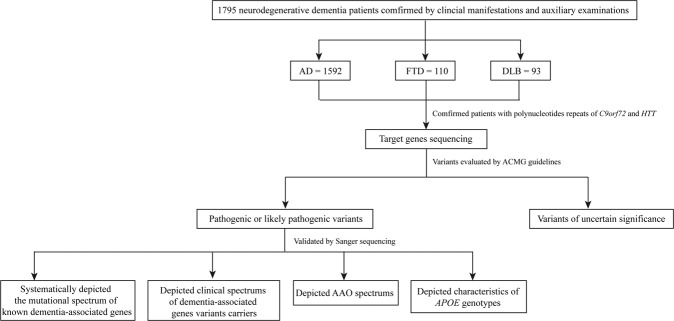
All P/LP variants were estimated by PCR amplification and Sanger sequencing using a Big Dye Terminator V3.1 on an ABI 3730xl DNA analyzer (Applied Biosystems, Foster City, USA). The DNA sequences were then analyzed using Sequencher software version 4.2. All primers were designed using Primer 5, and the primer sequences and PCR reaction conditions are listed in Supplementary Table 3. Meanwhile, variants of unknown significance identified in this study are shown in Supplementary Table 4. The study workflow is shown in Fig. 5.
Fig. 5. Workflow of this study.
AD Alzheimer’s disease, FTD frontotemporal dementia, DLB dementia with Lewy bodies.
Statistical analysis
Quantitative variables such as age at onset, age at diagnosis, disease duration, education attainment, and cognitive assessment score are expressed as the mean ± SD. All data were tested for normality and homogeneity of variance using the Shapiro-Wilk test and Levene variance equality test. Two independent samples were conducted using the t test or the Mann–Whitney U test. The χ2 test and Fisher exact test were used to analyze categorical data, such as the proportion of female patients, family history, the percentage of APOE4 positive or negative, and proportion of EOAD, LOAD or P/LP variants carriers and non-carriers patients. Multiple linear regression analysis was performed to correct the confounding factors and explore the factors affecting the AAO. All tests were two-tailed, and p < 0.05 was considered statistically significant. All analyses were performed using SPSS v.26 (IBM). Data were visualized using Prism 8 (GraphPad).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We are grateful to the participation of the patients and their family members in this study. This study was supported by the National Key R&D Program of China (Nos. 2020YFC2008500, 2017YFC0840100 and 2017YFC0840104 to L.S., No. 2018YFC1312003 to J.W.), the National Natural Science Foundation of China (Nos. 81671075 and 81971029 to L.S., No. 82071216 to B.J., and No. 81901171 to X.L.), Hunan Innovative Province Construction Project (No. 2019SK2335 to B.T.), and the Youth Science Foundation of Xiangya Hospital (No. 2018Q020 to X.L.).
Author contributions
B.J., H.L. and L.S. designed the study, interpreted the data, and draft the manuscript. L.G., X.X., X.L., Y.Z., L.W., Lu Z., X.W., Y.J, Q.Y. and Y.Z. participated in data acquisition and analysis. Lin Z., W.Z., J.W., X.Y., J.L., B.T. and L.S. performed the data analysis; All authors read and approved the final manuscript. B.J. and H.L. contributed equally to the manuscript.
Data availability
The sequencing raw data analyzed during this study has been deposited in European Variation Archive, the accession number was PRJEB46658. All other data are available from the corresponding authors on reasonable request.
Code availability
No custom code or scripts were used for the generation or processing of datasets.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Bin Jiao, Hui Liu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41525-021-00235-3.
References
- 1.Jack CR, et al. Prevalence of biologically vs clinically defined alzheimer spectrum entities using the national institute on aging–Alzheimer’s association research framework. JAMA Neurol. 2019;76:1174. doi: 10.1001/jamaneurol.2019.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alzheimer’s Association. 2020 Alzheimer’s disease facts and figures. Alzheimers Dement.16, 391–460 (2020).
- 3.Van Mossevelde S, Engelborghs S, van der Zee J, Van Broeckhoven C. Genotype-phenotype links in frontotemporal lobar degeneration. Nat. Rev. Neurol. 2018;14:363–378. doi: 10.1038/s41582-018-0009-8. [DOI] [PubMed] [Google Scholar]
- 4.Vann JS, O’Brien JT. The prevalence and incidence of dementia with Lewy bodies: a systematic review of population and clinical studies. Psychol. Med. 2014;44:673–683. doi: 10.1017/S0033291713000494. [DOI] [PubMed] [Google Scholar]
- 5.Hinz FI, Geschwind DH. Molecular genetics of neurodegenerative dementias. Cold Spring Harb. Perspect. Biol. 2017;9:a023705. doi: 10.1101/cshperspect.a023705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rademakers R, Neumann M, Mackenzie IR. Advances in understanding the molecular basis of frontotemporal dementia. Nat. Rev. Neurol. 2012;8:423–434. doi: 10.1038/nrneurol.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rohrer JD, et al. The heritability and genetics of frontotemporal lobar degeneration. Neurology. 2009;73:1451–1456. doi: 10.1212/WNL.0b013e3181bf997a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erkkinen, M. G., Kim, M. O. & Geschwind, M. D. Clinical neurology and epidemiology of the major neurodegenerative diseases. Cold Spring Harb. Perspect. Biol.10, a033118 (2018). [DOI] [PMC free article] [PubMed]
- 9.Guerreiro R, et al. Investigating the genetic architecture of dementia with Lewy bodies: a two-stage genome-wide association study. Lancet Neurol. 2018;17:64–74. doi: 10.1016/S1474-4422(17)30400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernardi L, et al. Novel PSEN1 and PGRN mutations in early-onset familial frontotemporal dementia. Neurobiol. Aging. 2009;30:1825–1833. doi: 10.1016/j.neurobiolaging.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Ishikawa A, et al. A mutantPSEN1 causes dementia with lewy bodies and variant Alzheimer’s disease. Ann. Neurol. 2005;57:429–434. doi: 10.1002/ana.20393. [DOI] [PubMed] [Google Scholar]
- 12.Geiger JT, et al. Next-generation sequencing reveals substantial genetic contribution to dementia with Lewy bodies. Neurobiol. Dis. 2016;94:55–62. doi: 10.1016/j.nbd.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raux, G. et al. Dementia with prominent frontotemporal features associated with L113P presenilin 1 mutation. Neurology55, 1577–1578 (2000). [DOI] [PubMed]
- 14.Kelley BJ, et al. Alzheimer disease-like phenotype associated with the c.154delA mutation in progranulin. Arch. Neurol. 2010;67:171–177. doi: 10.1001/archneurol.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coppola G, et al. Evidence for a role of the rare p.A152T variant in MAPT in increasing the risk for FTD-spectrum and Alzheimer’s diseases. Hum. Mol. Genet. 2012;21:3500–3512. doi: 10.1093/hmg/dds161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orme, T. et al. Analysis of neurodegenerative disease-causing genes in dementia with Lewy bodies. Acta Neuropathol. Commun.8, 5 (2020). [DOI] [PMC free article] [PubMed]
- 17.Mishra A, et al. Gene-based association studies report genetic links for clinical subtypes of frontotemporal dementia. Brain. 2017;140:1437–1446. doi: 10.1093/brain/awx066. [DOI] [PubMed] [Google Scholar]
- 18.Tsuang D, et al. APOE epsilon4 increases risk for dementia in pure synucleinopathies. JAMA Neurol. 2013;70:223–228. doi: 10.1001/jamaneurol.2013.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pasanen P, et al. A novel α-synuclein mutation A53E associated with atypical multiple system atrophy and Parkinson’s disease-type pathology. Neurobiol. Aging. 2014;35:2180.e1–2180.e5. doi: 10.1016/j.neurobiolaging.2014.03.024. [DOI] [PubMed] [Google Scholar]
- 20.Markopoulou K, et al. Clinical, neuropathological and genotypic variability in SNCA A53T familial Parkinson’s disease: Variability in familial Parkinson’s disease. Acta Neuropathol. 2008;116:25–35. doi: 10.1007/s00401-008-0372-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bougea A, et al. Frontotemporal dementia as the presenting phenotype of p.A53T mutation carriers in the alpha-synuclein gene. Parkinsonism Relat. Disord. 2017;35:82–87. doi: 10.1016/j.parkreldis.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 22.Ryan NS, et al. Clinical phenotype and genetic associations in autosomal dominant familial Alzheimer’s disease: a case series. Lancet Neurol. 2016;15:1326–1335. doi: 10.1016/S1474-4422(16)30193-4. [DOI] [PubMed] [Google Scholar]
- 23.Jia L, et al. PSEN1, PSEN2, and APP mutations in 404 Chinese pedigrees with familial Alzheimer’s disease. Alzheimers Dement. 2020;16:178–191. doi: 10.1002/alz.12005. [DOI] [PubMed] [Google Scholar]
- 24.Perez-Tur, J. et al. A further presenilin 1 mutation in the exon 8 cluster in familial Alzheimer’s disease. Neurodegeneration5, 207–212 (1996). [DOI] [PubMed]
- 25.Gómez-Isla, T. et al. A novel presenilin-1 mutation increased beta-amyloid and neurofibrillary changes. Ann. Neurol.41, 809–813 (1997). [DOI] [PubMed]
- 26.Kwok, J. B. J. et al. Presenilin-1 mutation L271V results in altered exon 8 splicing and Alzheimer’s disease with non-cored plaques and no neuritic dystrophy. J. Biol. Chem.278, 6748–6754 (2003). [DOI] [PubMed]
- 27.Bird TD, et al. Wide range in age of onset for chromosome 1—related familial Alzheimer’s disease. Ann. Neurol. 1996;40:932–936. doi: 10.1002/ana.410400619. [DOI] [PubMed] [Google Scholar]
- 28.Sherrington R. Alzheimer’s disease associated with mutations in presenilin 2 is rare and variably penetrant. Hum. Mol. Genet. 1996;5:985–988. doi: 10.1093/hmg/5.7.985. [DOI] [PubMed] [Google Scholar]
- 29.Jiang H, Jayadev S, Lardelli M, Newman M. A review of the familial Alzheimer’s disease locus PRESENILIN 2 and its relationship to PRESENILIN 1. J. Alzheimers Dis. 2018;66:1323–1339. doi: 10.3233/JAD-180656. [DOI] [PubMed] [Google Scholar]
- 30.Lin C, et al. Familial Alzheimer’s disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer’s disease type 3 gene. Nature. 1995;376:775–778. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- 31.Walker ES, Martinez M, Brunkan AL, Goate A. Presenilin 2 familial Alzheimer’s disease mutations result in partial loss of function and dramatic changes in Abeta 42/40 ratios. J. Neurochem. 2005;92:294–301. doi: 10.1111/j.1471-4159.2004.02858.x. [DOI] [PubMed] [Google Scholar]
- 32.Finckh U, et al. Variable expression of familial Alzheimer disease associated with presenilin 2 mutation M239I. Neurology. 2000;54:2006–2008. doi: 10.1212/WNL.54.10.2006. [DOI] [PubMed] [Google Scholar]
- 33.Giovagnoli AR, Marcon G, Giaccone G, Confaloni AM, Tagliavini F. Cognitive deficits in familial Alzheimer’s disease associated with M239V mutation of presenilin 2. Dement. Geriatr. Cogn. Disord. 2006;22:238–243. doi: 10.1159/000094972. [DOI] [PubMed] [Google Scholar]
- 34.Jiao B, et al. Mutational analysis in early-onset familial Alzheimer’s disease in Mainland China. Neurobiol. Aging. 2014;35:1957.e1–1957.e6. doi: 10.1016/j.neurobiolaging.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 35.De Jonghe C. Pathogenic APP mutations near the gamma-secretase cleavage site differentially affect Abeta secretion and APP C-terminal fragment stability. Hum. Mol. Genet. 2001;10:1665–1671. doi: 10.1093/hmg/10.16.1665. [DOI] [PubMed] [Google Scholar]
- 36.Murrell JR, Hake AM, Quaid KA, Farlow MR, Ghetti B. Early-onset Alzheimer disease caused by a new mutation (V717L) in the amyloid precursor protein gene. Arch. Neurol. 2000;57:885–887. doi: 10.1001/archneur.57.6.885. [DOI] [PubMed] [Google Scholar]
- 37.Ajroud-Driss S, et al. Mutation in the novel nuclear-encoded mitochondrial protein CHCHD10 in a family with autosomal dominant mitochondrial myopathy. Neurogenetics. 2015;16:1–9. doi: 10.1007/s10048-014-0421-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang M, et al. Mutation analysis ofCHCHD10 in different neurodegenerative diseases. Brain. 2015;138:e380–e380. doi: 10.1093/brain/awv082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bannwarth S, et al. A mitochondrial origin for frontotemporal dementia and amyotrophic lateral sclerosis through CHCHD10 involvement. Brain. 2014;137:2329–2345. doi: 10.1093/brain/awu138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Auranen M, et al. CHCHD10 variant p.(Gly66Val) causes axonal Charcot-Marie-Tooth disease. Neurol. Genet. 2015;1:e1. doi: 10.1212/NXG.0000000000000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao T, et al. Identification of CHCHD10 mutation in Chinese patients with Alzheimer disease. Mol. Neurobiol. 2016;54:5243–5247. doi: 10.1007/s12035-016-0056-3. [DOI] [PubMed] [Google Scholar]
- 42.Hara K, et al. Association of HTRA1 mutations and familial ischemic cerebral small-vessel disease. N. Engl. J. Med. 2009;360:1729–1739. doi: 10.1056/NEJMoa0801560. [DOI] [PubMed] [Google Scholar]
- 43.Verdura E, et al. Heterozygous HTRA1 mutations are associated with autosomal dominant cerebral small vessel disease. Brain. 2015;138:2347–2358. doi: 10.1093/brain/awv155. [DOI] [PubMed] [Google Scholar]
- 44.Nozaki H, et al. Distinct molecular mechanisms of HTRA1 mutants in manifesting heterozygotes with CARASIL. Neurology. 2016;86:1964–1974. doi: 10.1212/WNL.0000000000002694. [DOI] [PubMed] [Google Scholar]
- 45.Di Donato I, et al. Heterozygous mutations of HTRA1 gene in patients with familial cerebral small vessel disease. CNS Neurosci. Ther. 2017;23:759–765. doi: 10.1111/cns.12722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petkau TL, Leavitt BR. Progranulin in neurodegenerative disease. Trends Neurosci. 2014;37:388–398. doi: 10.1016/j.tins.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 47.Balendra R, Isaacs AM. C9orf72-mediated ALS and FTD: multiple pathways to disease. Nat. Rev. Neurol. 2018;14:544–558. doi: 10.1038/s41582-018-0047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poorkaj P, et al. Tau is a candidate gene for chromosome 17 frontotemporal dementia. Ann. Neurol. 1998;43:815–825. doi: 10.1002/ana.410430617. [DOI] [PubMed] [Google Scholar]
- 49.Burrell JR, Kiernan MC, Vucic S, Hodges JR. Motor Neuron dysfunction in frontotemporal dementia. Brain. 2011;134:2582–2594. doi: 10.1093/brain/awr195. [DOI] [PubMed] [Google Scholar]
- 50.Van Langenhove T, et al. Predicting development of amyotrophic lateral sclerosis in frontotemporal dementia. J. Alzheimer’s Dis. 2017;58:163–170. doi: 10.3233/JAD-161272. [DOI] [PubMed] [Google Scholar]
- 51.Lomen-Hoerth. C., Anderson, T. & Miller, B. The overlap of amyotrophic lateral sclerosis and frontotemporal dementia. Neurology59, 1077–1079 (2002). [DOI] [PubMed]
- 52.Ringholz GM, et al. Prevalence and patterns of cognitive impairment in sporadic ALS. Neurology. 2005;65:586–590. doi: 10.1212/01.wnl.0000172911.39167.b6. [DOI] [PubMed] [Google Scholar]
- 53.Watts GD, et al. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat. Genet. 2004;36:377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- 54.Al-Obeidi E, et al. Genotype-phenotype study in patients with valosin-containing protein mutations associated with multisystem proteinopathy. Clin. Genet. 2018;93:119–125. doi: 10.1111/cge.13095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blacker D, et al. ApoE-4 and age at onset of Alzheimer’s disease: the NIMH genetics initiative. Neurology. 1997;48:139–147. doi: 10.1212/WNL.48.1.139. [DOI] [PubMed] [Google Scholar]
- 56.Corder EH, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 57.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McKenna A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164–e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.den Dunnen JT, et al. HGVS recommendations for the description of sequence variants: 2016 update. Hum. Mutat. 2016;37:564–569. doi: 10.1002/humu.22981. [DOI] [PubMed] [Google Scholar]
- 61.DeJesus-Hernandez M, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bean L, Bayrak-Toydemir P. American College of Medical Genetics and Genomics Standards and Guidelines for Clinical Genetics Laboratories, 2014 edition: technical standards and guidelines for Huntington disease. Genet Med. 2014;16:e2–e2. doi: 10.1038/gim.2014.146. [DOI] [PubMed] [Google Scholar]
- 63.Hooli BV, et al. Rare autosomal copy number variations in early-onset familial Alzheimer’s disease. Mol. Psychiatry. 2014;19:676–681. doi: 10.1038/mp.2013.77. [DOI] [PubMed] [Google Scholar]
- 64.Alzheimer’s Disease Collaborative Group. The structure of the presenilin 1 (S182) gene and identification of six novel mutations in early onset AD families. Nat. Genet.11, 219–222 (1995). [DOI] [PubMed]
- 65.Qiu Q, et al. A novel PSEN1 M139L mutation found in a Chinese pedigree with early-onset Alzheimer’s disease increases Aβ42/Aβ40 ratio. J. Alzheimer Dis. 2019;69:199–212. doi: 10.3233/JAD-181291. [DOI] [PubMed] [Google Scholar]
- 66.Koriath, C. et al. Predictors for a dementia gene mutation based on gene-panel next-generation sequencing of a large dementia referral series. Mol Psychiatry.25, 3399–3412 (2020). [DOI] [PMC free article] [PubMed]
- 67.Jin SC, et al. Pooled-DNA sequencing identifies novel causative variants in PSEN1, GRN and MAPT in a clinical early-onset and familial Alzheimer’s disease Ibero-American cohort. Alzheimers Res. Ther. 2012;4:34–34. doi: 10.1186/alzrt137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Church A, et al. A novel presenilin 1 mutation, I202F occurring at a previously predicted pathogenic site causing autosomal dominant Alzheimer’s disease. Neurobiol. Aging. 2011;32:556.e1–556.e2. doi: 10.1016/j.neurobiolaging.2010.09.030. [DOI] [PubMed] [Google Scholar]
- 69.Raux G. Molecular diagnosis of autosomal dominant early onset Alzheimer’s disease: an update. J. Med. Genet. 2005;42:793–795. doi: 10.1136/jmg.2005.033456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Coleman P, Kurlan R, Crook R, Werner J, Hardy J. A new presenilin Alzheimer’s disease case confirms the helical alignment of pathogenic mutations in transmembrane domain 5. Neurosci. Lett. 2004;364:139–140. doi: 10.1016/j.neulet.2004.04.030. [DOI] [PubMed] [Google Scholar]
- 71.Houlden H, et al. A novel presenilin mutation (M233V) causing very early onset Alzheimer’s disease with Lewy bodies. Neurosci. Lett. 2001;313:93–95. doi: 10.1016/S0304-3940(01)02254-6. [DOI] [PubMed] [Google Scholar]
- 72.Campion D, et al. Mutations of the presenilin I gene in families with early-onset Alzheimer’s disease. Hum. Mol. Genet. 1995;4:2373–2377. doi: 10.1093/hmg/4.12.2373. [DOI] [PubMed] [Google Scholar]
- 73.Gomez-Isla T, et al. A novel presenilin-1 mutation: increased beta-amyloid and neurofibrillary changes. Ann. Neurol. 1997;41:809–813. doi: 10.1002/ana.410410618. [DOI] [PubMed] [Google Scholar]
- 74.Aldudo J, Bullido MJ, Arbizu T, Oliva R, Valdivieso F. Identification of a novel mutation (Leu282Arg) of the human presenilin 1 gene in Alzheimer’s disease. Neurosci. Lett. 1998;240:174–176. doi: 10.1016/S0304-3940(97)00950-6. [DOI] [PubMed] [Google Scholar]
- 75.Ikeda M, et al. The clinical phenotype of two missense mutations in the presenilin I gene in Japanese patients. Ann. Neurol. 1996;40:912–917. doi: 10.1002/ana.410400614. [DOI] [PubMed] [Google Scholar]
- 76.Campion D, et al. A large pedigree with early-onset Alzheimer’s disease: Clinical, neuropathologic, and genetic characterization. Neurology. 1995;45:80–85. doi: 10.1212/WNL.45.1.80. [DOI] [PubMed] [Google Scholar]
- 77.Rogaev EI, et al. Familial Alzheimer’s disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer’s disease type 3 gene. Nature. 1995;376:775–778. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 78.Ancolio K, et al. Unusual phenotypic alteration of beta amyloid precursor protein (betaAPP) maturation by a new Val-715 —> Met betaAPP-770 mutation responsible for probable early-onset Alzheimer’s disease. Proc. Natl Acad. Sci. USA. 1999;96:4119–4124. doi: 10.1073/pnas.96.7.4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Goate A. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature. 1991;6311:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 80.Delisle M, et al. A mutation at codon 279 (N279K) in exon 10 of the Tau gene causes a tauopathy with dementia and supranuclear palsy. Acta Neuropathol. 1999;98:62–77. doi: 10.1007/s004010051052. [DOI] [PubMed] [Google Scholar]
- 81.Guven G, et al. Mutation frequency of the major frontotemporal dementia genes, MAPT, GRN and C9ORF72 in a Turkish cohort of dementia patients. PLoS ONE. 2016;11:e0162592. doi: 10.1371/journal.pone.0162592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Piaceri I, et al. Novel GRN mutations in Alzheimer’s disease and frontotemporal lobar degeneration. J. Alzheimer Dis. 2018;62:1683–1689. doi: 10.3233/JAD-170989. [DOI] [PubMed] [Google Scholar]
- 83.Zhang W, Xie F, Lu P. Two novel heterozygous HTRA1 mutations in two pedigrees with cerebral small vessel disease families. Neurol. Sci. 2018;39:497–501. doi: 10.1007/s10072-017-3231-z. [DOI] [PubMed] [Google Scholar]
- 84.Bersano A, et al. Inclusion body myopathy and frontotemporal dementia caused by a novel VCP mutation. Neurobiol. Aging. 2009;30:752–758. doi: 10.1016/j.neurobiolaging.2007.08.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing raw data analyzed during this study has been deposited in European Variation Archive, the accession number was PRJEB46658. All other data are available from the corresponding authors on reasonable request.
No custom code or scripts were used for the generation or processing of datasets.