Fig. 5. 5MP represses RAN translation through eIF3c.
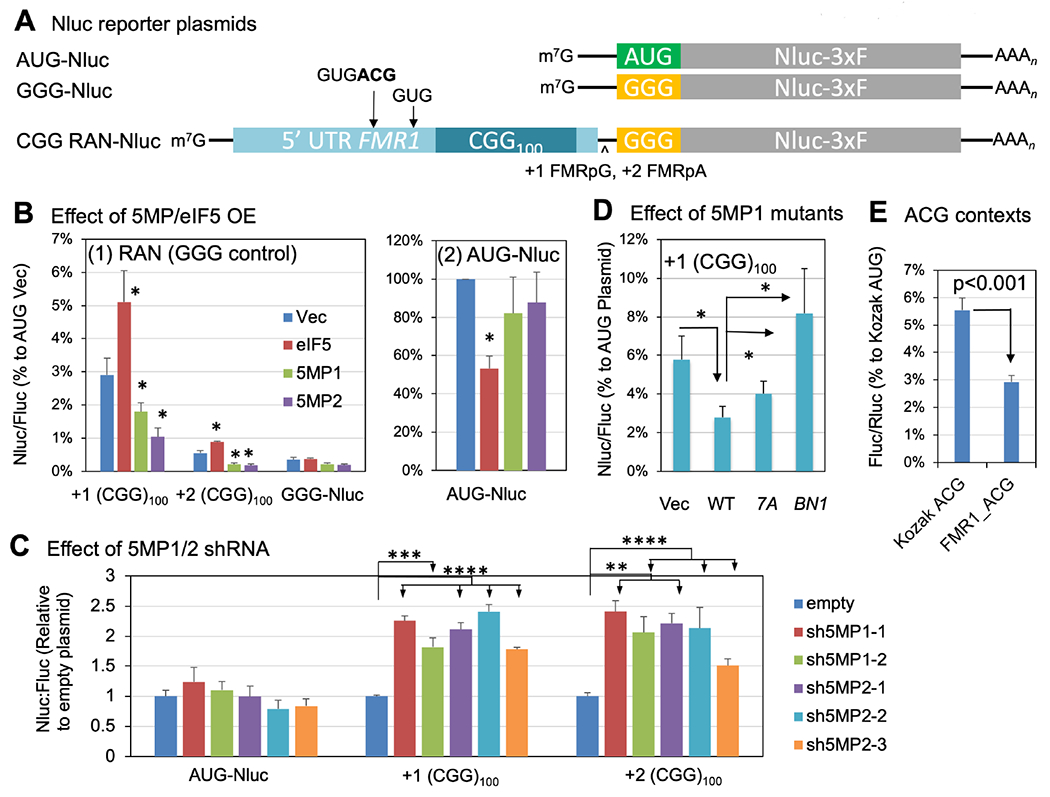
(A) NanoLuciferase (Nluc)-3xF plasmids used in this study. Arrows indicate the location of non-AUG start codon initiating translation of the +1 CGG reading frame (Kearse et al., 2016); the ACG codon studied in panel (E) was boldfaced. ^, 1 or 2 base-insertions to make FMR1-polyG or -polyA in frame to Nluc. (B) HEK293T transfected with indicated NanoLuciferase-3xF plasmid, a eIF5 or 5MP1/2-expressing plasmid and a firefly luciferase plasmid was assayed for nano- and firefly luciferase activities. Nluc/Fluc ratio was normalized to the values with AUG vector control transfectant. Bars indicate standard errors. *, p<0.05 compared to vector control in 4 or more independent experiments. (C) Expression of Nluc RAN translation and AUG Fluc control reporters coexpressed with empty plasmid or plasmid expressing sh5MPs (sh5MP1-1, p1462; sh5MP1-2, p1465; sh5MP2-1, p1467; sh5MP2-2, p1469; sh5MP2-3, p1470, see Key Resource Table) in HEK293Ts. Bars represent mean +/− SEM, N=6. 2 way ANOVA:p<0.0001. Two tailed Student’s T-test with Bonferroni correction, **p<0.01, ***p<0.001, ****p<0.0001. (D) Effect of 5MP1 mutations. Assays were done similar to (B) with 5MP1 plasmids with indicated mutations. Nluc/Fluc ratio was normalized to the value from AUG-Nluc transfectant with the same expression plasmid [e. g. the value from +1(CGG)100/5MP1 compared to that from AUG/5MP1] to correct for changes in AUG expression by each plasmid. *p<0.05 in 7 or more experiments. (E) ACG initiation frequency from its isolated FMR1 context located outside of the repeats. Firefly luciferase activity from the reporter bearing the 24-nt-long region upstream of the ACG codon was assessed along with a control Rluc reporter and compared to the values from Kozak AUG and ACG contexts (n=6), as in Fig. 3, after normalizing to one from Kozak AUG.
