Fig. 7. Model of translation initiation and its control by 5MP.
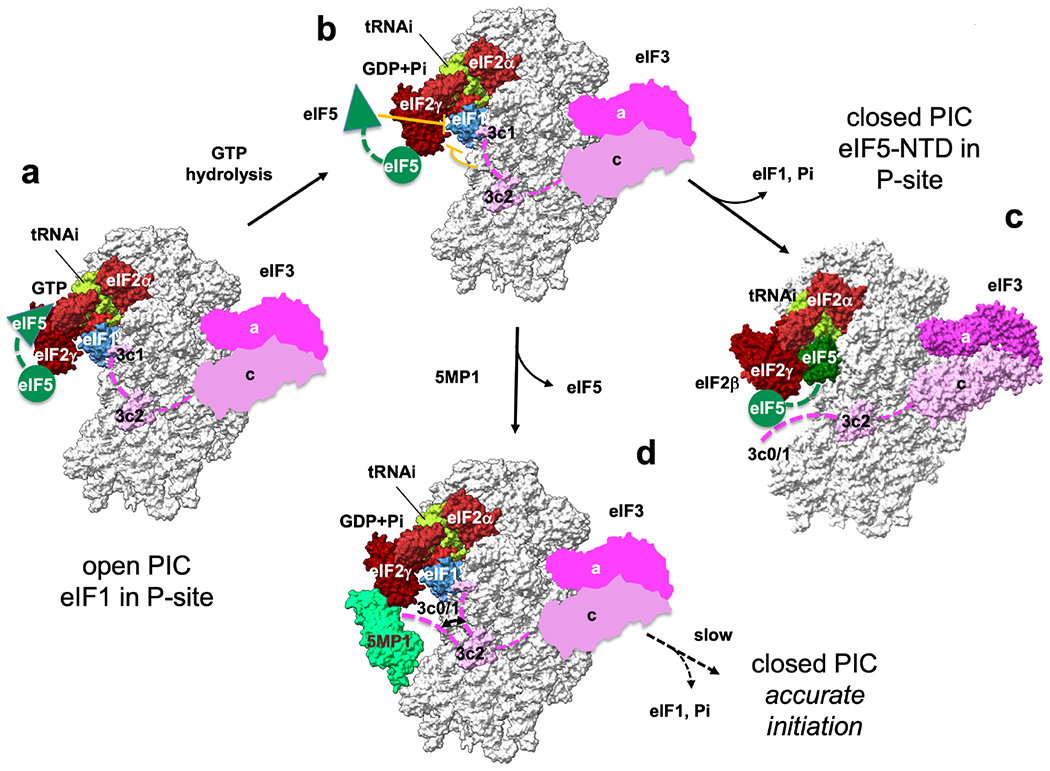
(a) MFC components stimulate a change to the scanning-competent open PIC conformation, while eIF5-NTD catalyzes GTP hydrolysis for eIF2. (b) During scanning, eIF1 plays the central role in keeping the open conformation necessary for accurate initiation at AUG codon. However, eIF5-NTD (Llacer et al., 2018) and eIF3c N-terminal tail termed eIF3c0 (Obayashi et al., 2017) intrinsically destabilize this conformation (stop bars in orange). For this reason, eukaryotes allow a certain level of general and repeat-associated non-AUG translation. (c) On AUG recognition, eIF1 and Pi are released to trigger the conformational change to the closed state, leading to translation initiation. eIF5-NTD plays a pivotal role in this process by physically excluding eIF1 from the P-site and binding to the P-site (Llacer et al., 2018). (d) 5MP can increase the accuracy of initiation by antagonizing the destabilizing effects caused by both eIF5-NTD and eIF3c0. Open and closed PIC structures are based on 3JAQ and 6FYX, respectively. In open PICs, the location of eIF3 is based on 6FYX. The location of eIF5-CTD (green circle) is based on (Bochler et al., 2020). Green triangle denotes eIF5-NTD responsible for GTP hydrolysis for eIF2 and docking into P-site on AUG recognition. Green dotted line, the linker between NTD and CTD of eIF5. Pink dotted line, eIF3c-N-terminal tail.
