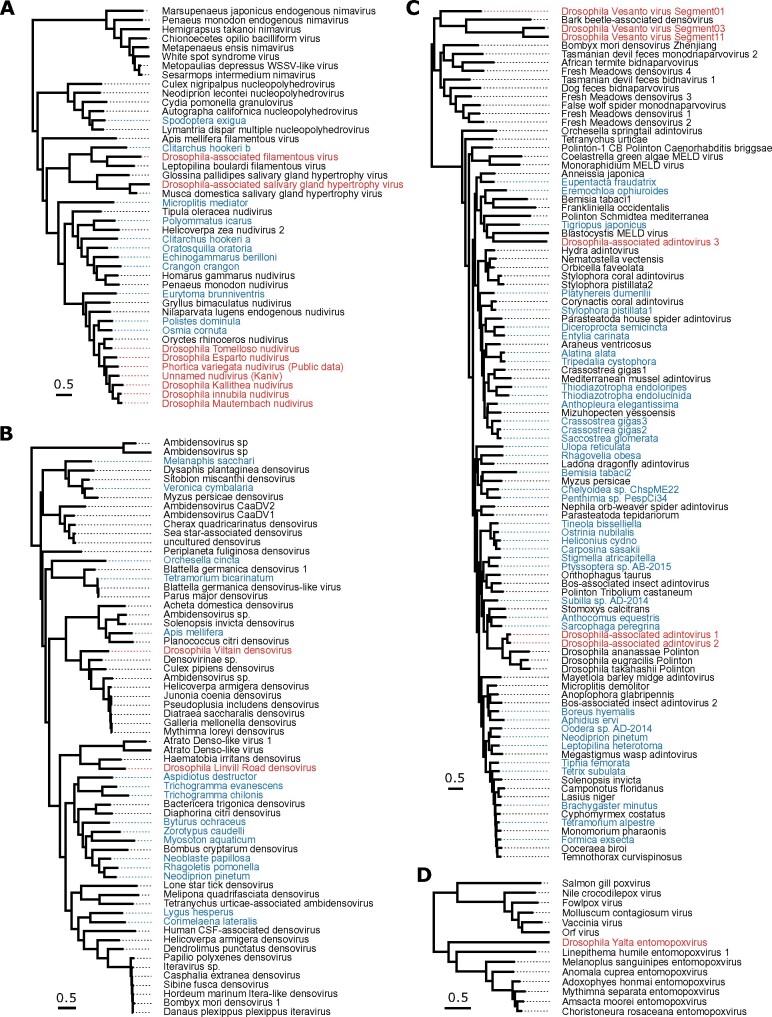
Figure 2.
Phylogenetic relationships. (A) Nudiviruses, hytrosaviruses, filamentous viruses, nucleopolyhedrosis viruses and nimaviruses, inferred from six concatenated protein coding genes. Note that these lineages are extremely divergent, and the alignment is not reliable at deeper levels of divergence. (B) Densoviruses, inferred from NS1. (C) Bidnaviruses (sometimes labelled ‘densovirus’) and adintoviruses (including representative polintons), inferred from DNA Polymerase B. (D) Pox and entomopoxviruses, inferred from three concatenated protein coding genes. All phylogenies were inferred from protein sequences by maximum likelihood, and scale bars represent 0.5 amino-acid substitutions per site. In each case, trees are mid-point rooted, viruses reported from Drosophila are shown in red, and sequences identified from virus transcripts in publicly available transcriptome assemblies are shown in blue, labelled by host species. The nudivirus from Phortica variegata was derived from PRJNA196337 (Vicoso and Bachtrog 2013). Alignments and tree files with bootstrap support are available in Supplementary Material.