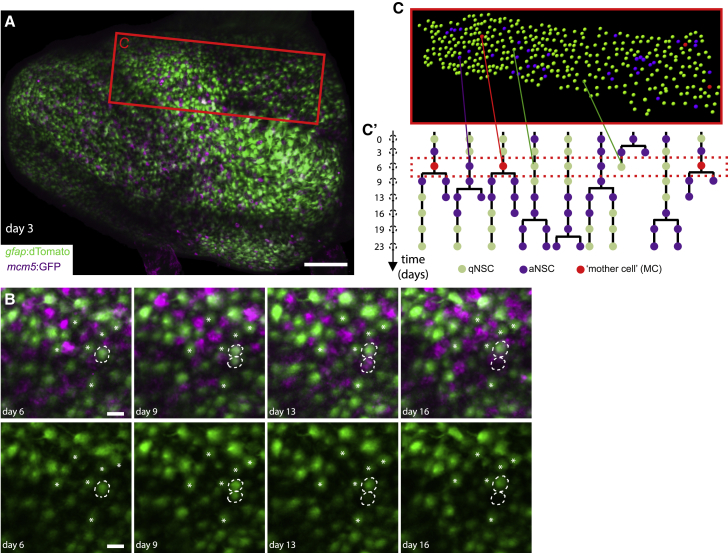
Figure 3.
Intravital imaging resolves adult NSC lineage trees in time and space
(A) Whole pallial hemisphere imaged intravitally in a 3mpf casper;Tg(gfap:dTomato);Tg(mcm5:GFP) fish (named Mimi) (anterior left, tile of 4 images with 10% overlap, taken at day 3 from a 35-day session of recordings every 3–4 days, 8 time points). Colors of the live reporters were adjusted (green: NSCs, magenta: proliferating cells).
(B) Close-ups from the same video showing an asymmetric NSC division (dotted circles) between days 3 and 9: 1 daughter differentiates over the next 7 days (bottom dotted circle, loss of the gfap:dTomato signal). White asterisks: random qNSCs close to the division, used for alignment.
(C and C’) Segmentation and NSC tracking over 23 days in Dm in Mimi. (C) Segmentation of ~390 cells per time point (area boxed in A, at day 6). (C’) Example of dividing tracks, with cell states (color-coded) and the spatial position of each tree (see also Figure S5A). Scale bars: (A), 100 μm; (B), 20 μm.