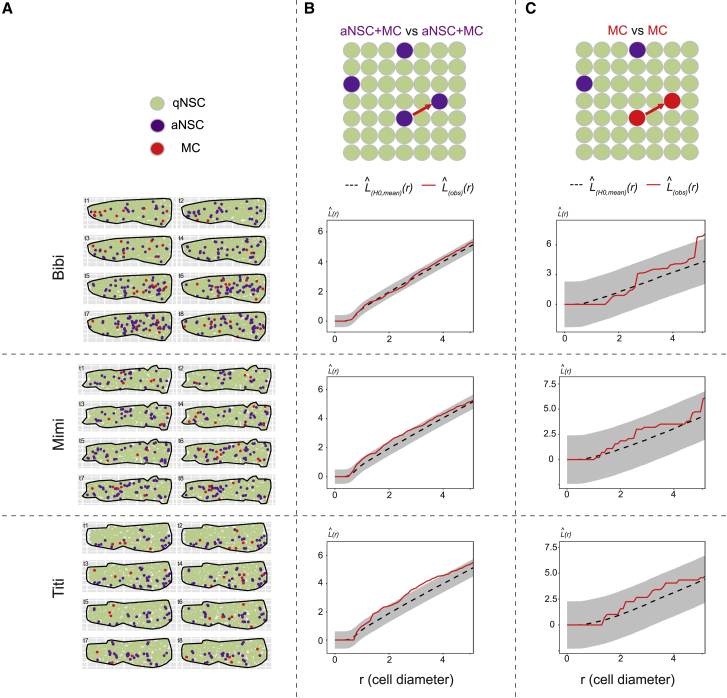
Figure 4.
Static spatial analysis of NSC activation from dynamic datasets
(A) Dm surfaces segmented for the 3 fish analyzed (Bibi, Mimi, Titi) at all time points, cell states color-coded (see also Figures S5B–S5F).
(B and C) L functions comparing the positions of aNSCs+MCs with one another (B), and MCs with one another (C) for each fish (see also Figures S5A–S5F). L(obs)(r) (red lines): experimental values, L functions for all time points pooled using a weighted average; L(H0, mean)(r) (black dotted lines): means under the random labeling null hypothesis; r: mean NSC diameter for each Dm surface; gray regions: 95% confidence envelopes. Pooled p > 0.01 in (B) and (C) for the [0–2] (r) interval.