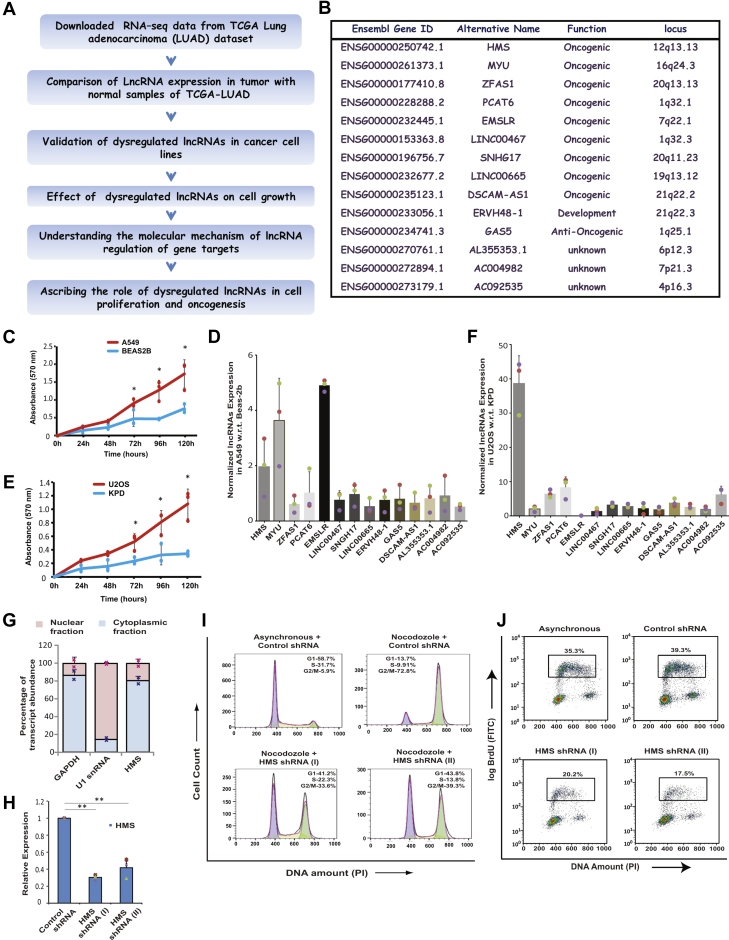
Figure 1.
Comparison of transcriptional profiles of TCGA cancer samples identifies dysregulated lncRNAs.A, schematic outline of the strategy employed for identification of dysregulated lncRNAs, their validation, identification of their gene targets and determining their effect on cell proliferation and oncogeneis. B, list of 14 selected lncRNAs. From the 224 upregulated lncRNAs, we selected 14 lncRNAs based on literature search, genomic screens of lncRNA function, proximity to cis-genes, and expression levels in various tissues. The table lists the Ensembl gene ID, alternate names, previously reported function and genomic locus. It should be noted that the screen identifies many lncRNAs across the genome that have been previously shown to have oncogenic activity. C, MTT proliferation assay to compare the growth rate of A549, an adenocarcinomic human alveolar epithelial cell line, with BEAS-2B, a nontumorigenic lung epithelial cell line, derived from a human lung tissue. The absorbance at 570 nm (minus the plate background absorbance at 630 nm) reflects the proportion of viable cells for each cell line at the indicated time interval. The data represents mean of three independent experiments ±standard deviation (S.D). Individual data points of the line chart have been shown as markers of the same colour. p-values calculated using two-tailed Student’s t test display that cell proliferation rate in A549 is significantly different from BEAS-2B samples at the indicated time intervals (∗p < 0.05). D, relative expression of 14 selected lncRNAs in A549 cells with respect to BEAS-2B cells, evaluated by individual quantitative real-time PCR. Beta-actin gene was used as the endogenous control for normalization of lncRNA expression in the two cell lines. The data represents mean of three independent experiments ±S.D. E, MTT proliferation assay to compare the growth rate of U2OS with KPD cell line as described in part C (n = 3, ∗p < 0.05, Student’s t test). F, evaluation of the levels of 14 lncRNAs in two human osteosarcoma cell: U2OS (rapid-proliferating, highly invasive) and KPD (slow-proliferating, less invasive) that have been previously used as contrasting pair of cell lines to study the role of ncRNAs in cancer. The data represents mean of three independent experiments ±S.D. G, subcellular fractionation of U2OS cells followed by evaluation of abundance of the indicated transcripts by qRT-PCR in nuclear and cytoplasmic fractions. The bar graph displays the percentage of the total amount of detected transcripts in different fractions. GAPDH and U1 snRNA serve as controls for cytoplasmic and nuclear fractions, respectively. The data is represented as mean ± SD from two independent experiments. Markers in pink and blue color point to individual data points of nuclear and cytoplasmic fractions, respectively. H, U2OS cells were transduced with lentiviral particles expressing either control shRNA or shRNAs targeting different regions of HMS: shRNA (I) and shRNA (II), followed by puromycin selection to obtain stable knockdown cells. The level of HMS was quantified by individual quantitative real-time PCR. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the endogenous control for normalization of HMS expression in different samples. The data represents mean of three independent experiments ±S.D. Note that both shRNA-I (∗∗p < 0.01, ANOVA/Tukey’s test) and shRNA-II (∗∗p < 0.01, ANOVA/Tukey’s test) significantly reduced HMS levels in comparison to control shRNA samples. I, depletion of HMS leads to an accumulation of cells in the G1 phase. U2OS cells stably depleted for HMS, as described in part H, were cultured in media with or without nocodazole for 16 h followed by cell cycle distribution analysis by flow cytometry. The inset shows the distribution of cells in different phases of the cell cycle. J, depletion of HMS impedes S phase progression. Flow cytometry of control or HMS-depleted U2OS cells, pulsed with BrdU. Dot plot displays BrdU incorporation (y-axis) and DNA content (x-axis). The inset shows the percentage of cells incorporating BrdU. Individual data points have been shown for all charts.