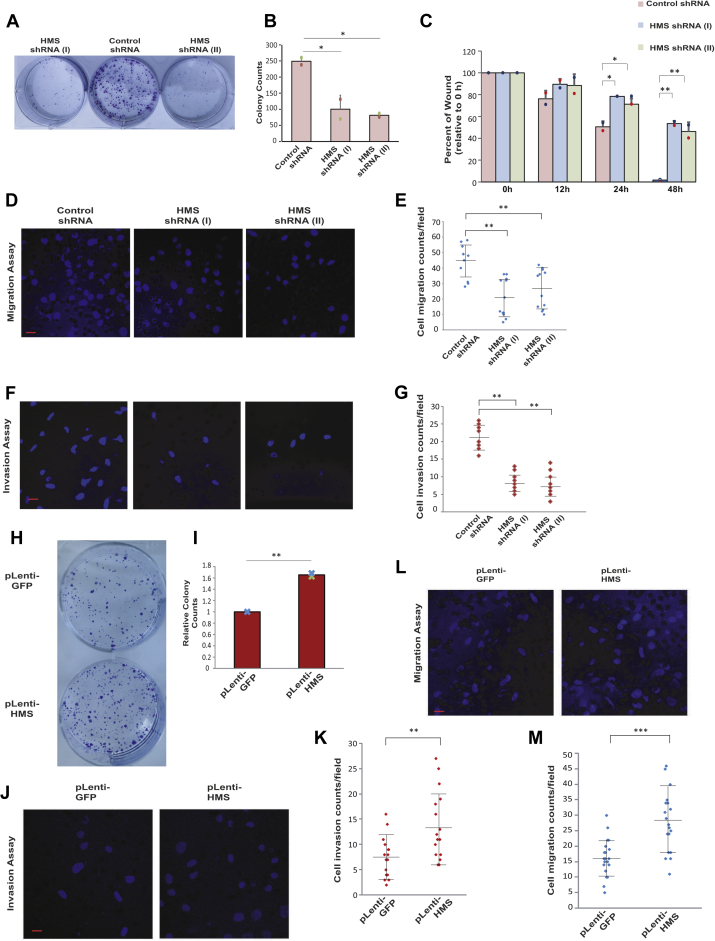
Figure 2.
HMS supports aggressive cancer-associated phenotypes.A, clonogenic assay to evaluate the effect of HMS depletion. HMS-depleted U2OS cells as described in Figure 1 were allowed to grow for 12 days, stained with crystal violet and the colonies were counted. B, quantification of number of colonies observed in part A. The data represents mean of two independent experiments ±S.D. Note that both shRNA-I (∗p < 0.05, ANOVA/Tukey’s test) and shRNA-II (∗p < 0.05, ANOVA/Tukey’s test) significantly reduced the colony forming ability in comparison to control shRNA samples. C, effect of HMS depletion on wound healing assay: HMS-depleted U2OS cells were grown to confluence after which a wound was created using a micropipette tip. The extent of wound healing was monitored at the indicated time points. The data represents mean of two independent experiments ±S.D. Note that in comparison to control shRNA samples, both shRNA-I (∗p < 0.05, ANOVA/Tukey’s test) and shRNA-II (∗p < 0.05, ANOVA/Tukey’s test) significantly reduced the wound healing capability at 24 h. Similarly, at 48 h shRNA-I (∗∗p < 0.01, ANOVA/Tukey’s test) and shRNA-II (∗∗p < 0.01, ANOVA/Tukey’s test) significantly reduced the wound healing capability. D, HMS depletion inhibits cell migration. A representative field showing the DAPI stained nucleus of control or HMS-depleted U2OS cells migrating through a microporous membrane counted 24 h after seeding. Scale bar, 20 μm. E, quantification of cell migration observed in part D. Each point refers to the number of cells in an individual field captured from two independent experiments, whereas long and short horizontal bars represent the mean and SD respectively of all fields. Note that in comparison to control shRNA samples, both shRNA-I (∗∗p < 0.01, ANOVA/Tukey’s test) and shRNA-II (∗∗p < 0.01, ANOVA/Tukey’s test) significantly reduced the cell migration. F, HMS depletion inhibits cell invasion. A representative field showing the DAPI stained nucleus of control or HMS-depleted U2OS cells invading through a Matrigel-coated membrane counted 24 h after seeding. Scale bar, 20 μm. G, quantification of cell invasion observed in part F. Each point refers to the number of cells in an individual field captured from two independent experiments, whereas long and short horizontal bars represent the mean and SD respectively of all fields. Note that in comparison to control shRNA samples, both shRNA-I (∗∗p < 0.01, ANOVA/Tukey’s test) and shRNA-II (∗∗p < 0.01, ANOVA/Tukey’s test) significantly reduced the cell invasion. H, clonogenic assay to evaluate the effect of HMS expression on cell proliferation. U2OS cells were transduced with lentiviral particles produced by transfection of pLenti-HMS or pLenti-GFP vector in HEK293T cells, followed by puromycin selection and generation of stable cells. U2OS cells stably expressing GFP or HMS were allowed to grow for 12 days, stained with crystal violet and the colonies were counted. I, quantification of number of colonies observed in part H. The data represents mean of two independent experiments ±S.D (∗∗p < 0.01, Student’s t test). J, HMS expression enhances cell invasion. A representative field showing the DAPI stained nucleus of U2OS cells, stably expressing GFP or HMS, as described in part H, invading through a Matrigel-coated membrane counted 24 h after seeding. Scale bar, 20 μm. K, quantification of cell invasion observed in part J. Each point refers to the number of cells in an individual field captured from two independent experiments, whereas long and short horizontal bars represent the mean and SD respectively (∗∗p < 0.01, Student’s t test). L, HMS expression enhances cell migration. A representative field showing the DAPI stained nucleus of U2OS cells, stably expressing GFP or HMS, as described in part H, migrating through a microporous membrane counted 24 h after seeding. Scale bar, 20 μm. M, quantification of cell migration observed in part L. Each point refers to the number of cells in an individual field captured from two independent experiments, whereas long and short horizontal bars represent the mean and SD respectively (∗∗∗p < 0.001, Student’s t test). Individual data points have been shown for all charts.