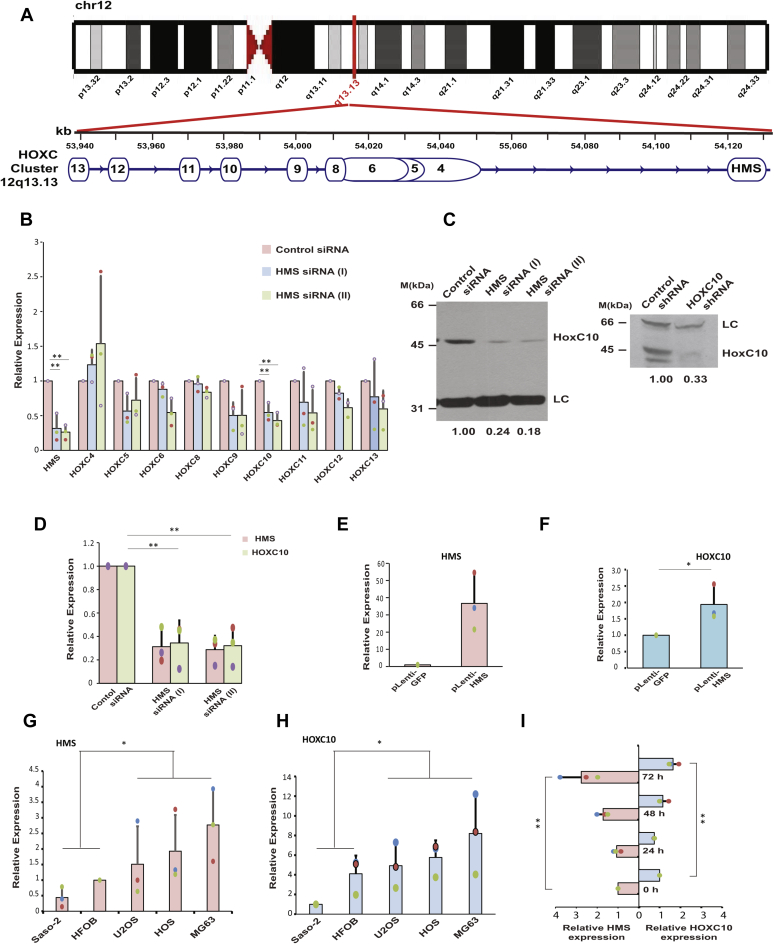
Figure 3.
HOXC10 gene, present 135 kb upstream of HMS locus, is downregulated in the absence of HMS.A, G-banded ideogram representing human chromosome 12, showing the cytogentic location of HOXC cluster and HMS gene at the 12q13.13 band. The genomic coordinates shown in kilobases (kb) are as per Genome Reference Consortium Human Build 38 patch release 13 (GRCh38.p13). The HOXC cluster constitutes of nine genes, namely HOXC13, HOXC12, HOXC11, HOXC10, HOXC9, HOXC8, HOXC6, HOXC5, and HOXC4. According to this assembly, the locus of HOXC10 gene is from 53985146 to 53990279 while the locus of HMS gene is from 54126071 to 54132843. The HOX Cluster as well as the HMS gene is expressed in the same direction from the “+” strand of chromosome 12, as depicted by arrows. B, expression of HOXC10 gene is reduced after HMS depletion. U2OS cells were transfected on three consecutive days with one of the siRNAs: control GL2, HMS siRNA (1), or HMS siRNA (2), and the level of genes in the HOXC cluster was quantified by individual quantitative real-time PCR. The bar graph indicates the levels of individual HOXC genes in HMS depleted samples relative to the control GL2 sample. GAPDH was used as the endogenous control for normalization of HOXC gene expression in different samples. The data represents mean of three independent experiments ±S.D. HMS and HOXC10 transcript levels were significantly reduced after transfection of shRNA (∗∗p < 0.01; ANOVA/Tukey’s test). C, downregulation of HOXC10 protein after HMS depletion. U2OS cells were transfected on three consecutive days with either control or HMS siRNA and the level of HOXC10 protein was determined by immunoblotting with anti-HOXC10 antibody. LC, loading control, a nonspecific band that displays equal protein load in each lane. The numbers indicate the level of HOXC10 protein relative to control siRNA transfected cells. C, right panel, immunoblotting of control shRNA or HOXC10 shRNA transfected U2OS cell lysate confirms the specificity of anti-HOXC10 antibody. The levels of endogenous HMS and HOXC10 mRNA in the same samples, as determined by quantitative real-time PCR, are shown in part D. The data represents mean of three independent experiments ±S.D. (∗∗p < 0.01; ANOVA/Tukey’s test). E, expression of HOXC10 gene is moderately increased after overexpression of HMS. U2OS cells were transduced with lentiviral particles produced by transfection of pLenti-HMS or pLenti-GFP vector in HEK293T cells, followed by puromycin selection and generation of stable cells. The cells were harvested 48 h after transduction followed by determination of HMS and HOXC10 mRNA levels by quantitative real-time PCR and shown in part E and F, respectively. The data has been represented as the mean ± SD of three independent experiments (∗p < 0.05, Student’s t test). G and H, relative expression of HMS and HOXC10 expression in different OS cell lines with respect to hFOB1.19, a human normal osteoblastic cell line. The expression of HMS (part G) in both nonaggressive cell lines, hFOB1.19 and Saos-2, is significantly different from the aggressive OS cell lines including U2OS, HOS, and MG63. The data has been represented as the mean ± SD of three independent experiments (∗p < 0.05, Student’s t test). The levels of HOXC10 shown in part H are significantly different between these two groups of cell lines. The data has been represented as the mean ± SD of three independent experiments (∗p < 0.05, Student’s t test). I, relative expression of HMS and HOXC10 in U2OS cells treated with 200 nM doxorubicin at the indicated time points. The levels of HMS and HOXC10 have been expressed w.r.t. to untreated cells. GAPDH was used as the endogenous control for normalization of HMS and HOXC10 expression in different samples. Note that the expression of HOXC10 increases concurrently with HMS. The data represents mean of three independent experiments ±S.D. Note that expression of both HOXC10 (∗∗p < 0.01, ANOVA/Tukey’s test) and HMS (∗∗p < 0.01, ANOVA/Tukey’s test) was significantly increased at 72 h after doxorubicin treatment. Individual data points have been shown for all charts.