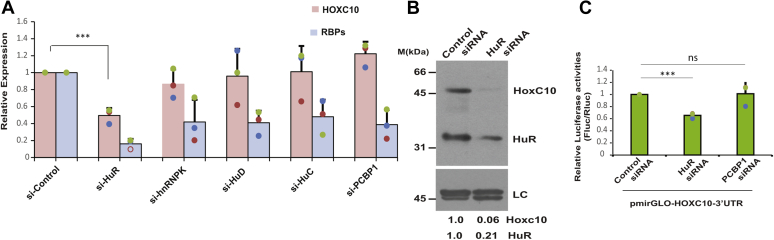
Figure 5.
HOXC10 3′UTR is destabilized in the absence of ELAV family protein, HuR.A, depletion of HuR downregulates HOXC10 mRNA. U2OS cells were transfected on three consecutive days with siRNA targeting individual RBPs (HuR; hnRNPK; HuD; HuC; PCBP1). The bar graph indicates the levels of specific RBPs and HOXC10 mRNA quantified by quantitative real-time PCR in different samples relative to control siRNA samples. GAPDH was used as the endogenous control for normalization of HOXC10 gene expression in different samples. The data has been represented as the mean ± SD of three independent experiments. Note that HOXC10 mRNA levels are significantly reduced after transfection of HuR siRNA (∗∗∗p < 0.0001, Student’s t test). B, downregulation of HOXC10 protein after HuR depletion. U2OS cells were transfected on three consecutive days with either control or HuR siRNA and levels of HOXC10 and HuR protein were determined by immunoblotting. LC, loading control, a nonspecific band that displays equal protein load in different lanes. The numbers indicate the levels of HOXC10 and HuR proteins relative to control siRNA transfected cells. C, HuR depletion downregulates the HOXC10 3′UTR-fused reporter luciferase activity. PmirGLO vector with the HOXC10 3′UTR cloned downstream of firefly luciferase ORF was transfected into U2OS cells depleted for HuR or PCBP1 and 24 h later luciferase activity was measured. The relative luciferase activity in each sample was expressed as a ratio of firefly to renilla luminescence. The data represents mean of three independent experiments ±S.D. Note that HuR siRNA-I significantly reduced the luciferase activity in comparison to control shRNA samples (∗∗∗p < 0.001, Student’s t test) while PCBP1 siRNA did not significantly alter the luciferase activity in comparison to control shRNA samples (ns, p = 0.9164, Student’s t test). Individual data points have been shown for all charts.