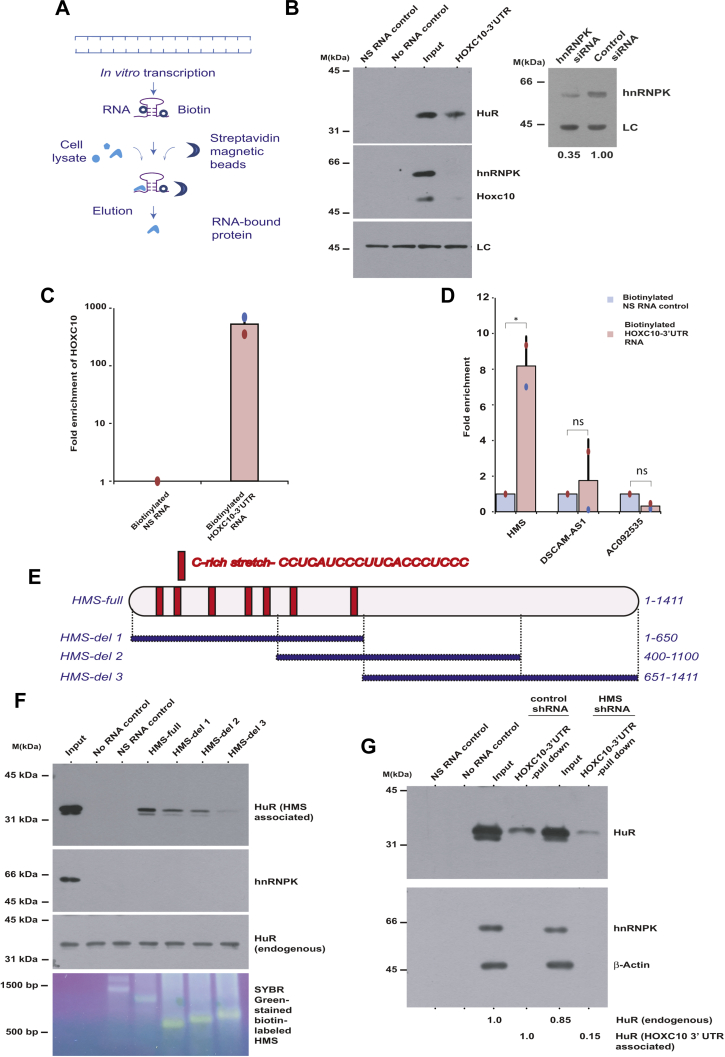
Figure 6.
HuR physically interacts with the cytosine-rich stretch of HMS to stabilize HOXC10 3′UTR.A, experimental design of RNA pull-down assay for identifying RNA-associated cellular proteins. RNA sequence was cloned in pCDNA3 plasmid downstream of the T7 RNA polymerase promoter followed by in vitro transcription in the presence of biotin-14-CTP to synthesize biotin-labeled RNA fragments. For RNA pull-down assay, the cell lysate was incubated with biotin-labeled RNA and streptavidin magnetic beads followed by washing, elution, and immunoblotting to detect the RNA-associated proteins. B, HuR physically associates with HOXC10 3′UTR. As per the protocol described in part A, in vitro synthesized biotinylated HOXC10 3′UTR RNA was incubated with U2OS cell lysates to identify proteins associated with biotinylated HOXC10 3′UTR. NS control is a nonspecific commercial biotin-labeled in vitro transcribed RNA while “no-RNA” serves as the negative control for nonspecific binding. Input lane shows 10% of total cell lysate used for the pull-down assay. LC displays that equal cell lysate was loaded in each assay. Note that hnRNP-K does not bind to HOXC10 3′UTR. B, right panel, immunoblotting of control GL2 or hnRNP-K siRNA transfected U2OS cell lysate confirms the specificity of anti-hnRNP-K antibody. C and D, RNA pull-down assay to identify the lncRNAs associated with biotinylated-HOXC10 3′UTR. Biotinylated-HOXC10 3′UTR was used for the precipitation of RNA from U2OS cell lysates and qRT-PCR was performed to identify the associated lncRNAs, HMS, DSCAM-AS1, and AC092535. Part C displays that biotinylated HOXC10 3′UTR bound effectively to the streptavidin beads, while Part D shows that HMS was specifically enriched with HOXC10 3′UTR bound beads. The data represents mean of two independent experiments ±S.D. Note that HMS associates with HOXC10 3′UTR in comparison to nonspecific RNA control (∗p < 0.05, Student’s t test) while AC092535 and DSCAM-AS1 (ns, p = 0.689, Student’s t test) do not significantly associate with HOXC10 3′UTR. E, schematic representation of HMS displaying its seven cytosine-rich stretches. The sequence of one of the cytosine-rich stretches has been shown at the top. The illustration shows the full-length and deletion fragments of HMS used for the precipitation of RNA-binding proteins from U2OS cell lysates. F, HuR physically associates with cytosine-rich stretches of HMS. Full-length HMS or its deletion fragments, as indicated in part E, were cloned in pCDNA3 downstream of the T7 RNA polymerase promoter followed by in vitro transcription in the presence of biotin-14-CTP to synthesize biotin-labeled RNA fragments. For RNA pull-down assay 1 μg of biotinylated full-length HMS or deletion fragments, as indicated, were used for the precipitation of HuR from U2OS cells and immunoblotting was performed to identify the associated HuR protein (top panel). SYBR-Green stained denaturing agarose gel displays that equal amounts of biotin-labeled full length HMS or deletion fragments were used for RNA pull-down assay (third panel). G, the physical association between HuR and HOXC10 3′UTR is disrupted in the absence of HMS. As described in part B, biotinylated HOXC10 3′UTR was used for the precipitation of HuR from U2OS cells transfected with either HMS or control GL2 siRNA. The input lane in GL2 and HMS siRNA samples shows that the endogenous HuR protein levels are not significantly decreased after HMS RNAi (0.85 fold in comparison to GL2 samples). However, the HuR associated with HOXC10 3′UTR is significantly decreased after HMS depletion (0.15 fold in comparison to GL2 samples). The amount of HuR associated with HOXC10 3′UTR was normalized to the levels of endogenous HuR in GL2 and HMS siRNA samples. Individual data points have been shown for all charts.