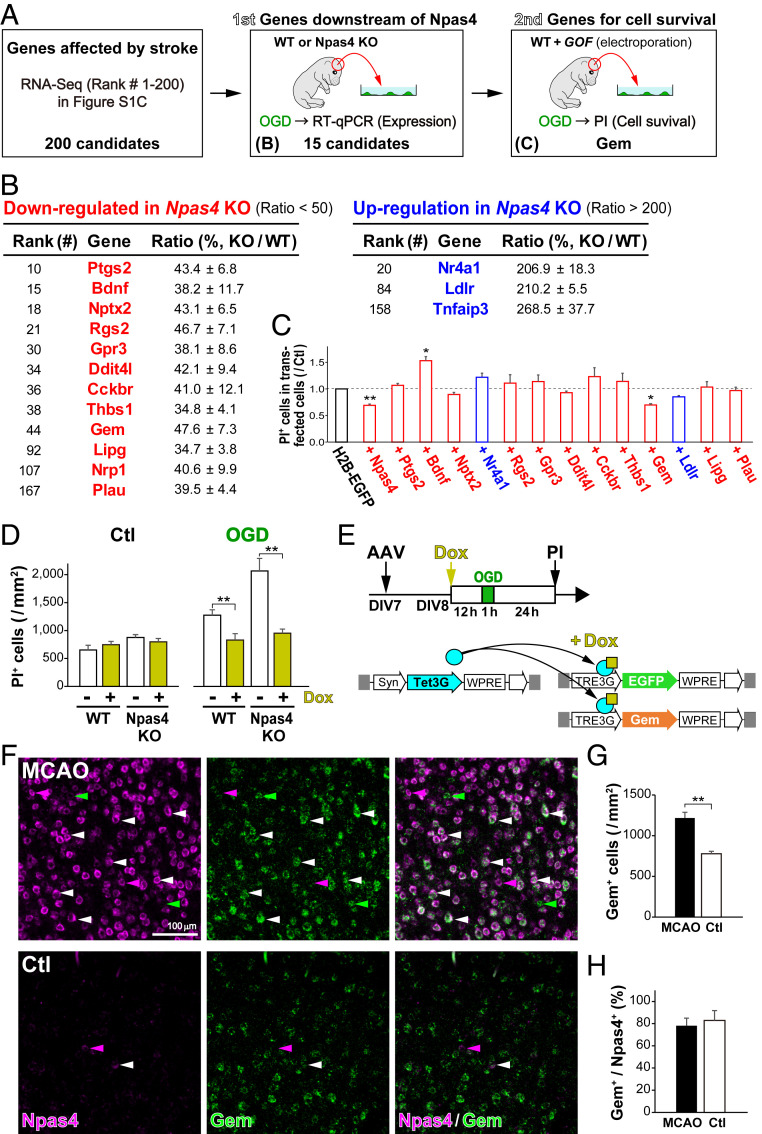
Fig. 5.
Search for Npas4 target genes conferring protection against ischemic death. (A) Schematic drawing for the search for Npas4 target genes in vitro. Genes whose expression was altered after MCAO were identified by RNA-Seq. (B) The first screen for candidate genes that were regulated in ischemic neurons in an Npas4-dependent manner. Among the top 200 genes listed in SI Appendix, Fig. S2C, expression of the 15 candidate genes was confirmed by qRT-PCR on primary cultured neurons from WT (n = 4) and Npas4 KO (n = 4) pregnant mice subjected to 1 h of OGD. (C) The second screen for the 13 differentially regulated genes listed in B. Primary neurons were cotransfected with each candidate gene and H2B-EGFP, and numbers of dead cells after OGD were counted via PI staining 24 h later. (D) Reduced numbers of dead cells overexpressing Gem in WT and Npas4-KO primary neurons after OGD. (E) Schematic drawings of Gem overexpression with the AAV/Syn–TetOn system in primary neurons and the experimental timeline. Primary neurons from WT and Npas4 KO embryonic cortexes were cotransfected with AAV/Syn–Tet3G and AAV/TRE3G–EGFP or AAV/TRE3G–Gem. After 12 h of Dox administration, primary neurons received 1 h of OGD and were subjected to PI staining 24 h later to count dead cells (D). Note that overexpression of Gem in Npas4-deficient neurons was sufficient to reduce the number of dead cells after ischemia. (F) Gem expression in cortical neurons after stroke. Two-color ISH with Npas4 (magenta) and Gem (green) was performed in the neocortex 1 h after MCAO. (G and H) The numbers of Gem+ cells (G) and the ratio of Gem+ cells to Npas4+ cells (H) in the MCAO side (MCAO) compared with those in the control side (Ctl). **P < 0.01 (Student’s t test in G and H, Student’s t test with Bonferroni correction in C and G, and two-way ANOVA with post hoc Tukey’s test in D).