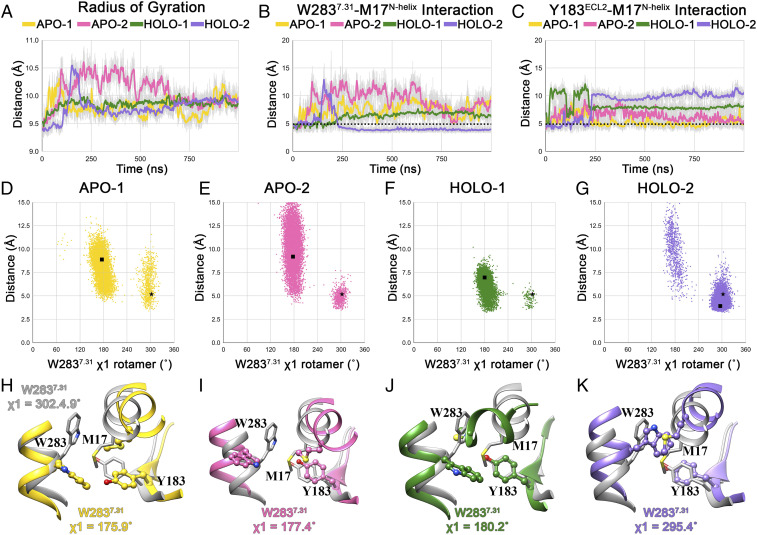
Fig. 6.
Conformational dynamics of the ligand-binding pocket in MD simulations. (A) Evolution of radius of gyration (50-point moving average) of the binding site residues in 1-µs trajectories. Evolution of S-π–type W2837.31–M17N-helix (B) and Y183ECL2–M17N-helix (C) interactions in 1-µs trajectories. Scatter plot showing distribution of W2837.31 (ring centroid)–M17N-helix (S-atom) distances and W2837.31 χ1 rotamer angles in W2837.31 apo-1 (D), apo-2 (E), holo-1 (F), and holo-2 (G) trajectories. Structures with W2837.31 χ1 rotamer angle equal to the average W2837.31 χ1 rotamer angle in the last 500 ns of the simulation are selected as representatives from each trajectory. The distance and χ1 rotamer angle corresponding to the crystal structure and representative structure from each simulation are shown using a black star and square in the scatter plots. Organization of the W2837.31–M17N-helix–Y183ECL2 bridge motif in representative structures, along with their W2837.31 χ1 rotamer angle, from apo-1 (H) (yellow), apo-2 (I) (pink), holo-1 (J) (green), and holo-2 (K) (purple) trajectories. The bridge motif from the crystal structure (gray) is also shown superposed to the representative structure.