Significance
Group 2 innate lymphoid cells (ILC2s) and adaptive CD4+ T helper type 2 (Th2) cells express a common effector program orchestrated by the “master” transcription factor GATA3 that is acquired through development or differentiation, respectively. To elucidate the regulatory mechanisms controlling the acquisition of this shared program, we used a combination of chromatin accessibility data and CRISPR/Cas9-mediated deletion, which revealed a Gata3 enhancer necessary for ILC2 development and function. Notably, this enhancer was largely dispensable for Th2 cell differentiation. Thus, ILC2s and Th2 cells display different requirements for the induction of a common type 2 helper effector program.
Keywords: ILC2, innate lymphoid cell, Th2 cell, type 2 immunity, GATA3
Abstract
The type 2 helper effector program is driven by the master transcription factor GATA3 and can be expressed by subsets of both innate lymphoid cells (ILCs) and adaptive CD4+ T helper (Th) cells. While ILC2s and Th2 cells acquire their type 2 differentiation program under very different contexts, the distinct regulatory mechanisms governing this common program are only partially understood. Here we show that the differentiation of ILC2s, and their concomitant high level of GATA3 expression, are controlled by a Gata3 enhancer, Gata3 +674/762, that plays only a minimal role in Th2 cell differentiation. Mice lacking this enhancer exhibited defects in several but not all type 2 inflammatory responses, depending on the respective degree of ILC2 and Th2 cell involvement. Our study provides molecular insights into the different gene regulatory pathways leading to the acquisition of the GATA3-driven type 2 helper effector program in innate and adaptive lymphocytes.
Innate lymphoid cells (ILCs) are classically considered tissue-resident lymphocytes that are functionally divided into three groups (groups 1, 2, and 3), mirroring CD4+ T helper (Th) cell subsets, in accordance with their transcription factor (TF) expression profile and rapid cytokine response under varied homeostatic and inflammatory conditions (1). ILCs and T cells express a highly overlapping subset of TFs, yet these two lineages differ in their temporal acquisition of effector properties through development or activation, respectively (2, 3). Among the shared TFs expressed in both lineages, GATA3 (encoded by Gata3) in particular plays a central role. GATA3 is well established as an essential regulator of both ILC and T cell development, differentiation, and function (4, 5). In T cells, GATA3 controls the development of early thymic progenitors (ETPs) and thymopoiesis (6, 7); CD4–CD8 T cell lineage commitment (8); T cell homeostasis (9); and Th2 cell differentiation and function (10–12). Akin to its functions in T cells, GATA3 regulates ILC precursor development (13, 14), peripheral ILC maintenance (14, 15), ILC2 differentiation and function (16, 17), and may contribute to ILC vs. lymphoid tissue–inducer (LTi) lineage specification (18, 19). Importantly, GATA3 expression dramatically increases above developmental levels during type 2 lymphocyte lineage commitment in both ILC2s and Th2 cells (10, 18, 20, 21). GATA3 up-regulation promotes ILC2 and Th2 cell differentiation by activating type 2 helper effector genes (e.g., Il4/Il5/Il13 and Il33ra) and suppressing alternate lineage promoting factors (e.g., Tbx21 and Ifng) (4). How exactly the expression of Gata3 is regulated at multiple distinct stages within the ILC and T cell lineages remains poorly understood.
Proper tissue- and stage-specific regulation of gene expression is achieved partly through the combined interaction of cis-regulatory elements both proximal (promoters) and distal (i.e., enhancers). GATA3 is critically expressed in a variety of tissue contexts apart from the immune system. Gata3-null mice exhibit embryonic lethality (22), and tissue-specific enhancers regulate GATA3 expression during the development of the craniofacial ganglia (23), central nervous system, endocardium, urogenital system (24), kidney (25), inner ear (26), and embryonic lens (27). Of particular importance to the immune system, Engel and coworkers identified a 7.1-kb T cell–specific Gata3 enhancer (TCE7.1, Gata3 +278/285), which resides ∼280 kb downstream of Gata3 within a 2.5-Mb gene desert (21, 28). Through a combination of bacterial artificial chromosome and CRISPR/Cas9-mediated deletion approaches, Ohmura et al. demonstrated that a core 1.2-kb element of TCE7.1, Gata3 +283/284, was necessary for optimal Gata3 expression in ETPs, CD4 single positive (CD4SP) thymocytes, and naïve CD4+ T cells (28). However, no element of Gata3 +278/285 fully recapitulated the elevated pattern of Gata3 expression seen in Th2 cells (21, 28). Furthermore, the contribution of Gata3 +278/285 or other regulatory elements to Gata3 expression throughout the ILC lineage remained unexplored. While it is well established that the Gata3-mediated type 2 helper effector program of ILC2s and Th2 cells is acquired in very different contexts, i.e., development vs. immune challenge, respectively (2, 3), it is unclear whether these differences are reflected in the use of or reliance on discrete Gata3 regulatory elements. This question is of fundamental interest not only to understand the evolution of the type 2 helper effector program in these lineages, but also to develop potential strategies to selectively dissect and manipulate the innate and adaptive arms of type 2 immunity. For example, whether an innate type 2 response by ILC2s is a prerequisite for the effective differentiation of Th2 cells remains controversial (3, 29–31) and could be addressed using mice lacking an ILC2-specific enhancer.
Through a comparison of accessible chromatin regions within the Gata3 locus gene desert, we identified a regulatory region 674-kb downstream of the Gata3 gene that functions as a type 2–specific Gata3 enhancer (Gata3 +674/762). Deletion of this cis-regulatory region via CRISPR/Cas9 selectively impaired the differentiation and function of ILC2s at homeostasis, as well as during type 2 inflammation. This enhancer was highly selective for ILC2s and only partially and mostly indirectly impacted Th2 cells, resulting in variable functional defects in allergic and helminthic inflammatory responses, depending on the degree of requirement for ILC2s vs. Th2 cells. Other lymphoid lineages such as group 1 (natural killer [NK] cell and ILC1) and group 3 (NCR+ ILC3 and CD4+ LTi) ILCs as well as undifferentiated CD4+ T cells remained unperturbed. In contrast, we found that the previously identified enhancer Gata3 +278/285 functioned mainly during the early development of ILCs but not in the differentiation or function of ILC2s, indicating distinct control of these processes. Finally, application of an in vivo enhancer reporter strategy in conjunction with the analysis of chromatin accessibility profiles revealed that elements of Gata3 +674/762 and Gata3 +278/285 are available and active at distinct stages of Gata3 expression during ILC and T cell development, differentiation, and function.
Results
A Distal Enhancer of Gata3 Regulates ILC2 Homeostasis.
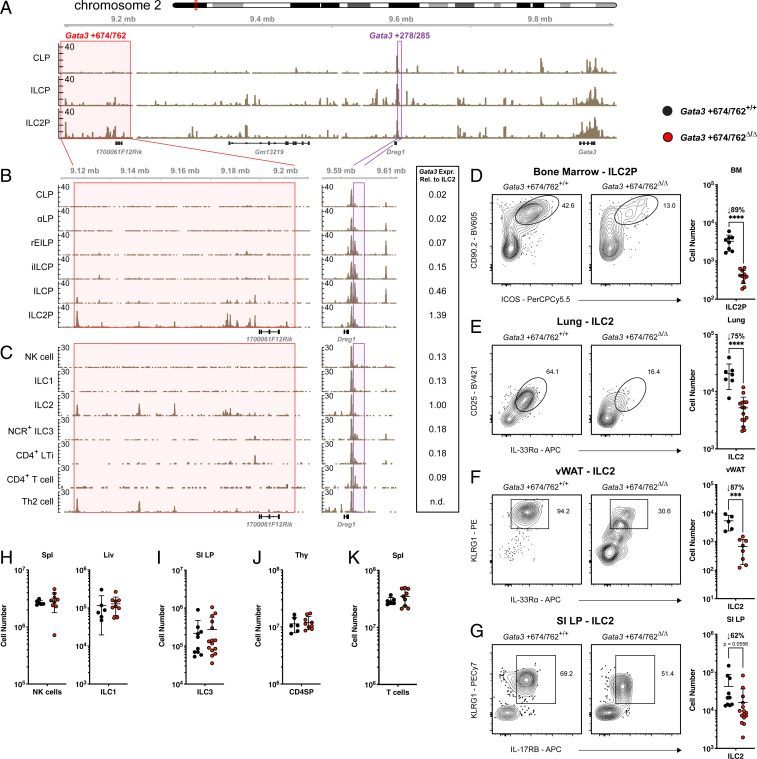
To identify potential cis-regulatory elements of Gata3 associated with ILC2 development we examined chromatin accessibility in bone marrow (BM) ILC progenitors using the assay for transposase-accessible chromatin with sequencing (ATAC-seq) (Fig. 1A). Approximately 674-kb downstream of Gata3 we identified an 88-kb region (Gata3 +674/762) containing signatures of type 2–specific activity. Chromatin accessibility increased in this region from the common lymphoid progenitor (CLP) to the ILC2 precursor (ILC2P), coinciding with ILC2 lineage differentiation. Moreover, within the 88-kb locus, an uncharacterized long noncoding RNA (lncRNA) 1700061F12Rik was expressed specifically in peripheral ILC2s, as determined by RNA-seq data from Immgen (SI Appendix, Fig. S1A) (32). Cell type–specific transcription of lncRNAs has been increasingly recognized as a correlate of active cis-regulatory elements. For example, the lncRNA Rroid was previously found to demarcate a group 1 ILC-specific enhancer of Id2 (33). Similarly, the Gata3 enhancer TCE7.1 (Gata3 +278/285) resides adjacent to the lncRNA Dreg1, which was coexpressed with Gata3 in T cells (Fig. 1A) (34, 35). A more detailed examination of accessibility within sequential progenitors throughout BM ILC development revealed that elements within the Gata3 +674/762 region began to show increased accessibility in the ILC precursor (ILCP), the stage in which ILC1/2/3 multilineage priming occurs, and were further augmented in the ILC2P where Gata3 expression is maximal (Fig. 1B, red window and side bar) (18, 36). In contrast, there was no detectable accessibility at earlier stages such as the refined early innate lymphoid precursor (rEILP) and the incipient ILC precursor (iILCP) where Gata3 expression was low (Fig. 1B, side bar). These results stood in contrast to accessibility at the previously identified enhancer Gata3 +278/285, which was accessible from the rEILP to the ILCP, but closed upon ILC2 differentiation (Fig. 1B, purple window).
Fig. 1.
Chromatin accessibility at the Gata3 locus and impact of Gata3 +674/762 deletion on lymphocyte subsets. (A) ATAC-seq accessibility coverage tracks in BM CLP, ILCP, and ILC2P. Red and purple windows represent Gata3 +674/762 and Gata3 +278/285 regions, respectively. (B) Zoomed in ATAC-seq accessibility coverage tracks for the Gata3 +674/762 and Gata3 +278/285 regions in sequential BM progenitors to the ILC lineage, including CLP, αLP (α4β7 lymphoid precursor), rEILP, iILCP, ILCP, and ILC2P. (C) Published ATAC-seq tracks of splenic NK cells, liver ILC1s, lung ILC2s, SI LP NCR+ ILC3s, SI LP CD4+ LTis, splenic CD4+ T cells, and N. brasiliensis–activated lung Th2 cells (2). Expression levels of Gata3-Citrine relative to lung ILC2s are shown to the Right of B and C (18). Representative flow cytometry plots and summary data of cell numbers in WT vs. Gata3 +674/762Δ/Δ mice for (D) BM ILC2P (pregated on Lin−α4β7+IL-7Rα+), (E) lung ILC2s (pregated on CD45.2+CD19−CD11c−CD3ε−TCRβ−IL-7Rα+CD90.2+), (F) vWAT ILC2s (pregated on CD45.2+CD19−CD11c−CD3ε−TCRβ−IL-7Rα+CD90.2+CD25+), and SI LP ILC2s (pregated on CD45.2+CD19−CD11c−CD3ε−TCRβ−IL-7Rα+CD90.2+Sca-1+). Summary data of cell numbers for (H) spleen NK cells and liver ILC1s, (I) SI LP ILC3s, (J) CD4SP thymocytes, and (K) splenic CD4+ T cells in WT vs. Gata3 +674/762Δ/Δ mice. Dots represent individual mice; n ranging from 6 to 15 in different groups pooled from multiple independent experiments; data are presented as mean ± SEM. Statistical comparison was performed via unpaired t test. ***P < 0.001; ****P < 0.0001.
Given that Gata3 is expressed throughout T cell development in the thymus and in mature ILCs and differentiated T cells in the periphery, we next evaluated chromatin accessibility in these populations using publicly available ATAC-seq data (2, 37). The distal Gata3 +674/762 region was conspicuously inaccessible in thymocytes, while Gata3 +278/285 accessibility persisted throughout T cell development, consistent with previously reported experiments (SI Appendix, Fig. S1B) (21, 28, 37). Among peripheral mature ILCs and T cells, only ILC2s and lung Th2 cells from Nippostrongylus brasiliensis–infected mice displayed sections of open chromatin within the Gata3 +674/762 locus, coinciding with differentiation and enhanced Gata3 expression (Fig. 1C, red window and side bar) (2, 10, 18). A comparison of accessibility across the Gata3 locus revealed no ILC2- vs. Th2 cell–specific peaks (SI Appendix, Fig. S2). At the Gata3 +278/285 enhancer, however, only group 3 ILCs and CD4+ T cells maintained some minor accessibility while this enhancer was closed in group 1 ILCs, ILC2s, and Th2 cells, despite continuous Gata3 expression throughout all populations (Fig. 1C, purple window and side bar). These results suggest that chromatin accessibility within the distal Gata3 +674/762 region is regulated in a distinct manner from Gata3 +278/285 in both ILCs and T cells, with opening of elements in Gata3 +674/762 coinciding with the acquisition and expression of a type 2 helper effector program by developing ILC2s and differentiating Th2 cells.
To directly evaluate the role of the Gata3 +674/762 region in vivo, we employed a CRISPR/Cas9-mediated deletion strategy to generate mice lacking this locus and evaluated representative populations (SI Appendix, Fig. S1C). Strikingly, Gata3 +674/762Δ/Δ mice displayed markedly diminished numbers of group 2 ILCs at homeostasis. ILC2P in the adult BM as well as mature ILC2 in the lung, visceral white adipose tissue (vWAT), and small intestinal lamina propria (SI LP) were reduced 62 to 89% compared to wild-type (WT) littermate controls (Fig. 1 D–G). Conversely, group 1 ILCs (NK cells and/or ILC1s) in the spleen, liver, lung, and vWAT as well as ILC3s in the SI LP were preserved in the absence of the Gata3 +674/762 region (Fig. 1 H and I and SI Appendix, Fig. S1D). Similarly, thymocytes, splenic T cells, and splenic B cells were also unaltered in the absence of Gata3 +674/762 (Fig. 1 J and K and SI Appendix, Fig. S1 E and F). Taken together, these results indicate a specific reliance on the Gata3 +674/762 region in ILC2s, which coincides with regional chromatin accessibility and heightened Gata3 expression, that was not observed in other Gata3-expressing cells at homeostasis. Hereafter we will refer to the deleted 88-kb region as the type 2–specific Gata3 enhancer (Gata3 +674/762).
ILC2s Bearing a Deletion of Gata3 +674/762 Are Functionally Impaired at Homeostasis.
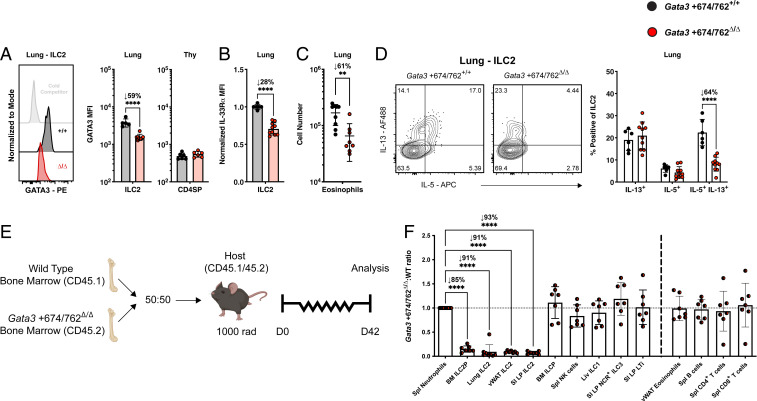
We next evaluated the differentiation and function of residual ILC2s at homeostasis in mice lacking the Gata3 +674/762 region. We found that Gata3 +674/762Δ/Δ mature ILC2s expressed 50 to 60% less GATA3 than WT littermate ILC2s in the lung and SI LP, while SI LP ILC3s, CD4SP thymocytes, and ETPs showed no reduction in GATA3 expression (Fig. 2A and SI Appendix, Fig. S1G). As GATA3 expression was significantly diminished in Gata3 +674/762Δ/Δ ILC2s, we then examined the expression of several GATA3 target genes. The surface receptor IL-33Rα is expressed by ILC2s in peripheral tissues, such as the lung and vWAT, and its expression is regulated by GATA3 in both ILC2s and Th2 cells (14, 38, 39). Relative to WT littermate controls, lung and vWAT ILC2s from Gata3 +674/762Δ/Δ mice displayed 28 to 33% lower levels of surface IL-33Rα (Fig. 2B and SI Appendix, Fig. S1H). GATA3 binds to several cis-regulatory elements within the Il4/Il5/Il13 cytokine locus in ILC2s and Th2 cells to control locus accessibility and gene expression (14, 17, 40–43). Basal production of IL-5 by ILC2s underlies the homeostatic recruitment and maintenance of eosinophils in several tissues (44, 45). In both the lung and vWAT, eosinophil numbers were reduced 61 to 79% in Gata3 +674/762Δ/Δ mice compared to WT littermate controls (Fig. 2C and SI Appendix, Fig. S1I). Diminished numbers of eosinophils could result from a combined effect of reduced ILC2 numbers and an impaired capacity to sense local IL-33; therefore, we addressed the intrinsic capacity of ILC2s to produce type 2 cytokines ex vivo. ILC2s haploinsufficient for GATA3 produce less IL-5 and IL-13 than their WT counterparts (17). Indeed, ILC2s from the lung and SI LP of Gata3 +674/762Δ/Δ mice, which expressed ∼50% less GATA3, contained 64% fewer IL-5/IL-13 double producers when stimulated ex vivo with phorbol myristate acetate (PMA) and ionomycin (Fig. 2D and SI Appendix, Fig. S1J). In sum, the aforementioned results indicate that deletion of the Gata3 +674/762 locus massively and globally impairs the frequency, differentiation, and function of ILC2s at homeostasis.
Fig. 2.
Impact of Gata3 +674/762 deletion on homeostatic ILC2 function. (A) Representative histogram and summary bar graph of GATA3 median fluorescent intensity (MFI) in lung ILC2s and summary bar graph for CD4SP thymocytes. White histogram denotes lung ILC2 GATA3 stain blocked with unlabeled antibody (cold competitor). (B) Summary bar graph of normalized IL-33Rα MFI on lung ILC2s. (C) Summary data of lung eosinophil cell numbers. (D) Representative flow cytometry plots and summary data for frequency of IL-5 and IL-13 cytokine production from lung ILC2s (pregated on CD45.2+CD19−CD11c−CD3ε−TCRβ−IL-7Rα+CD90.2+CD25+IL-33Rα+) stimulated in vitro with PMA and ionomycin for 4 h. (E) Schematic for establishing congenically marked mixed bone marrow chimeric mice. (F) Gata3 +674/762Δ/Δ:WT reconstitution ratio for the indicated populations in mixed bone marrow chimeric mice. Dots represent individual mice; n ranging from 6 to 10 in different groups pooled from multiple independent experiments; data are presented as mean ± SEM. Statistical comparison was performed via unpaired t test, multiple unpaired t test, or one-way ANOVA. **P < 0.01; ****P < 0.0001.
To ascertain the functional deficiency of Gata3 +674/762Δ/Δ ILC2s in vivo, we applied a 3-d regimen of type 2 pulmonary inflammation via intranasal (i.n.) administration of IL-33 (SI Appendix, Fig. S1K) (46, 47). Using this model, we found that eosinophilia was reduced in the lung and bronchoalveolar lavage (BAL) in Gata3 +674/762Δ/Δ mice, and infiltration of ILC2s into the bronchoalveolar space did not reach levels seen in WT littermate mice (SI Appendix, Fig. S1L). Thus, the inflammatory response to i.n. IL-33 administration is impaired in Gata3 +674/762Δ/Δ mice, likely a combined result of diminished ILC2 numbers and function.
To determine whether the observed reduction in ILC2s in Gata3 +674/762Δ/Δ mice was cell intrinsic, we used congenically marked mice to generate mixed bone marrow chimeras containing a 50:50 ratio of Gata3 +674/762Δ/Δ and WT bone marrow (Fig. 2E). Following 6 wk of reconstitution, ILC2s of Gata3 +674/762Δ/Δ origin were outcompeted 1:7 in the BM, 1:11 in the lung and vWAT, and 1:14 in the SI LP by WT ILC2s (Fig. 2F). In contrast, groups 1 and 3 ILCs as well as eosinophils, B cells, and T cells showed comparable reconstitution between Gata3 +674/762Δ/Δ and WT cells. From these results, we conclude that the Gata3 +674/762 region is intrinsically required to achieve normal numbers of ILC2s.
A subset of regulatory T (Treg) cells normally present within the adipose tissue express a type 2–like program characterized by the expression of GATA3 and IL-33Rα (44, 48, 49). At homeostasis, we observed a 77% reduction in GATA3+ Treg cells from Gata3 +674/762Δ/Δ mice compared to WT littermate controls (SI Appendix, Fig. S1M). Furthermore, this defect appeared to be cell extrinsic, as it was corrected in mixed (Gata3 +674/762Δ/Δ:WT) bone marrow chimeras (SI Appendix, Fig. S1N). Given that vWAT GATA3+ Treg cells proliferate in response to IL-33 (49), and adventitial stromal cells produce IL-33 in response to IL-13 from ILC2s (50), the homeostatic defect in the vWAT may be secondary to the ILC2 defect in Gata3 +674/762Δ/Δ mice.
Gata3 +278/285 Regulates Pan-ILC Development before Differentiation.
The previously identified enhancer, Gata3 +278/285, was shown to control the development and persistence of T cells at homeostasis; however, the contribution of Gata3 +278/285 to Gata3 expression within the ILC lineage was not explored (21, 28). To characterize the impact of Gata3 +278/285 on ILC development and function, we generated mice deficient in Gata3 +278/285 via CRISPR/Cas9-mediated deletion (SI Appendix, Fig. S3A). Similar to Gata3 +674/762Δ/Δ mice, Gata3 +278/285Δ/Δ mice showed a dramatic reduction in the number of ILC2P in the BM and ILC2 in the lung and SI LP compared to WT littermate controls (SI Appendix, Fig. S3 B–D). However, in contrast with Gata3 +674/762Δ/Δ, Gata3 +278/285 deletion also had a broad impact on other ILCs. In the spleen, liver, and lung of Gata3 +278/285Δ/Δ mice, group 1 ILCs were diminished, while ILC3s were overrepresented in the SI LP, possibly a result of an expanded niche from the loss of SI LP ILC2s (SI Appendix, Fig. S3 E and F) (51). Consistent with initial observations in Gata3 +278/285Δ/Δ mice, CD4SP T cells were considerably underrepresented in the spleen and in the thymus, causing an inversion of the CD4/CD8 ratio (SI Appendix, Fig. S3 G and H) (28). Splenic B cell numbers were unaffected by the deletion of Gata3 +278/285Δ/Δ (SI Appendix, Fig. S3I). Taken together, these results indicate a general reliance on Gata3 +278/285 for the development or homeostatic maintenance of GATA3-expressing ILCs and CD4+ T cells.
We next assessed the impact of Gata3 +278/285 deletion on the expression of GATA3 and GATA3 target genes. In striking contrast with Gata3 +674/762Δ/Δ mice, the residual mature ILC2s in the lung and SI LP of Gata3 +278/285Δ/Δ mice expressed unaltered levels of GATA3 compared to WT littermate ILC2s (SI Appendix, Fig. S3J). Conversely, GATA3 expression in ETPs and CD4SP thymocytes was diminished in Gata3 +278/285Δ/Δ mice, in line with a previous publication (28). Further contrasting observations made in Gata3 +674/762Δ/Δ mice, IL-33Rα expression on the residual Gata3 +278/285Δ/Δ lung ILC2s, and expression of IL-5 and IL-13 from stimulated Gata3 +278/285Δ/Δ lung and SI LP ILC2s, were also unaltered compared with that of WT littermate ILC2s (SI Appendix, Fig. S3 K and L). Collectively, these results demonstrate a differential impact for Gata3 +278/285 and Gata3 +674/762, with the former contributing more globally to the low/medium level of GATA3 expression during the development of ILCs and T cells and the latter specifically regulating elevated levels of GATA3 expression and acquisition of the type 2 helper effector program in ILC2s. Notably these findings correlate with the temporal pattern of accessibility of these enhancers and the relative expression of Gata3 in the different lymphocyte subsets (Fig. 1 B and C and SI Appendix, Fig. S1B) (18).
Gata3 +674/762Δ/Δ Mice Have an Impaired Type 2 Inflammatory Response.
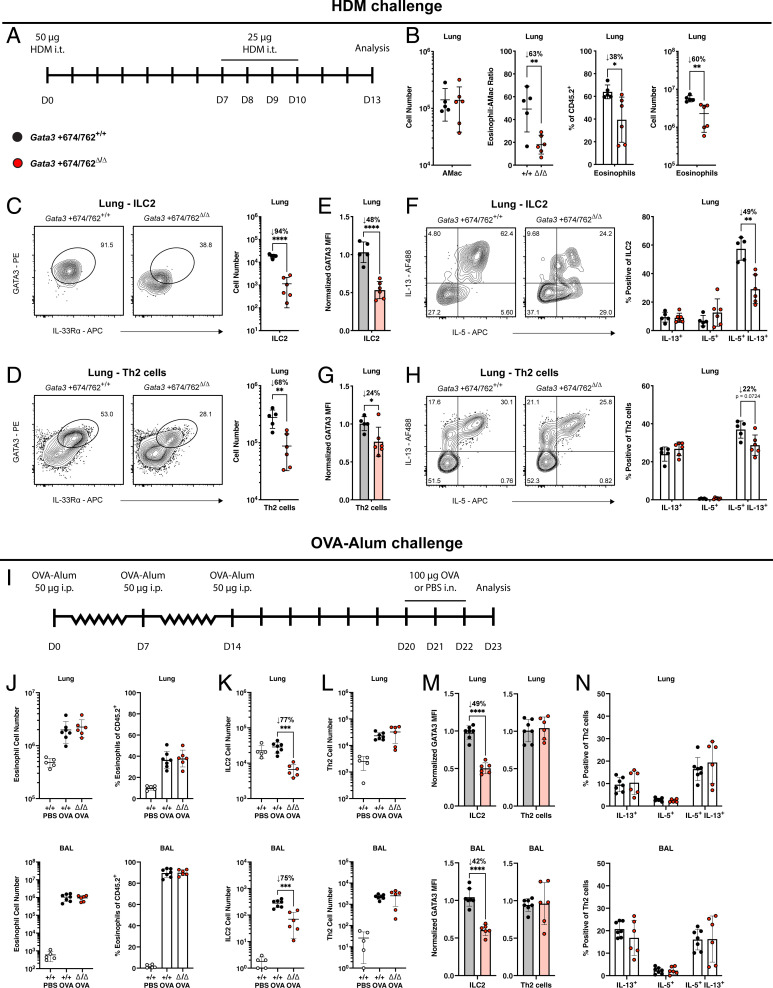
Th2 cells, like ILC2s, up-regulate GATA3 above their undifferentiated CD4+ counterparts (10, 21), and may similarly depend on Gata3 +674/762 for this enhanced expression. Rapid challenge with IL-33 in the absence of protein antigen precludes any significant contribution of Th2 cells to type 2 pulmonary inflammation (46). To broadly assess the impact of Gata3 +674/762 deletion in vivo, we used two experimental models of type 2 airway inflammation, which elicit both ILC2 and Th2 cell responses. First, we applied a 14-d protocol of intratracheal (i.t.) allergen sensitization and challenge using house dust mite (HDM) extract (Fig. 3A) (47). Following HDM challenge on days 7 to 10, Gata3 +674/762Δ/Δ mice exhibited 60 to 78% diminished eosinophilia in the lung and BAL, while maintaining a 94 to 96% reduction in ILC2 numbers (Fig. 3 B and C and SI Appendix, Fig. S4 A and B). Unlike studies at homeostasis, this model also permitted the assessment of Th2 cell generation and function, demonstrating a 68 to 86% impaired Th2 cell differentiation and infiltration in both the lung and BAL compared to WT littermate controls (Fig. 3D and SI Appendix, Fig. S4C). Furthermore, like ILC2s, GATA3 expression and the frequency of IL-5/IL-13 double producers were lower in Gata3 +674/762Δ/Δ Th2 cells (Fig. 3 E–H and SI Appendix, Fig. S4 D and E). Notably, however, the degree of reduction in each of these parameters was much less pronounced in Th2 cells compared to ILC2s: e.g., 94% vs. 68% for cell number, 48% vs. 24% for GATA3 expression, and 49% vs. 22% for IL-5/IL-13 double producers in lung ILC2s vs. lung Th2 cells, respectively. Thus, while ILC2s were drastically impaired in both number and function, relatively large numbers of Th2 cells were preserved with near normal function. Together these results explain why hallmarks of allergic inflammation were only partially compromised, including a 60% decrease in lung eosinophilia (Fig. 3B), and unaltered bronchial goblet cell hyperplasia (SI Appendix, Fig. S4F).
Fig. 3.
Impact of Gata3 +674/762 deletion on allergic airway inflammation. (A) Schematic for i.t. sensitization and challenge with HDM extract. (B) Representative flow cytometry plot and summary data for alveolar macrophages (AMac) and eosinophils in the lung of HDM-challenged mice. Representative flow cytometry plots and summary data of (C) ILC2 (pregated on CD45.2+CD19−CD11c−CD3ε−TCRβ−IL-7Rα+CD90.2+CD25+) and (D) Th2 cell (pregated on CD45.2+CD3ε+TCRβ+CD90.2+CD4+) numbers in the lung. (E) Summary plots of GATA3 MFI in lung ILC2s. (F) Representative flow cytometry plots and summary data for frequency of IL-5 and IL-13 cytokine production from lung ILC2s (pregated on CD45.2+CD19−CD11c−CD3ε−TCRβ−IL-7Rα+CD90.2+CD25+IL-33Rα+) stimulated in vitro with PMA and ionomycin for 4 h. (G) Summary plots of GATA3 MFI in lung Th2 cells. (H) Representative flow cytometry plots and summary data for frequency of IL-5 and IL-13 cytokine production from lung Th2 cells (pregated on CD45.2+CD3ε+TCRβ+CD90.2+CD4+IL-33Rα+) stimulated in vitro with PMA and ionomycin for 4 h. (I) Schematic of the OVA–alum model for induction of allergic airway inflammation via i.p. immunization with OVA–alum and subsequent i.n. challenge with OVA. (J) Summary data for eosinophils in the lung and BAL of OVA–alum-challenged mice. Summary data of (K) ILC2 and (L) Th2 cell numbers in the lung and BAL. (M) Summary plots of GATA3 MFI in ILC2s and Th2 cells from the lung and BAL. (N) Summary data for frequency of IL-5 and IL-13 cytokine production from lung Th2 cells stimulated in vitro with PMA and ionomycin for 4 h. Dots represent individual mice; n = 5 or 6 (HDM) and n = 7 or 6 (OVA–alum) for WT or Gata3 +674/762Δ/Δ mice. Data are pooled from multiple independent experiments and presented as mean ± SEM. Values of 0 were converted to a value of 1 on a log scale. Statistical comparison was performed via unpaired t test or multiple unpaired t test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
We confirmed the conclusions made in the HDM challenge model by employing a second model of type 2 inflammation using the helminth Strongyloides venezuelensis. From the site of subcutaneous (s.c.) infection, S. venezuelensis migrates through the lung where it induces a type 2 inflammatory response via activation of ILC2s and Th2 cells before eventual maturation in the duodenum of the small intestine and clearance within ∼2 wk in mice with a functional adaptive immune system (31, 52, 53). Subsequent to the resolution of helminth infection, Gata3 +674/762Δ/Δ mice showed weaker eosinophilia (63 to 84%); decreased numbers of ILC2s (84 to 94%) with lower expression of GATA3 (71 to 77%) and dual production of IL-5/IL-13 (17 to 57%); and blunted Th2 cell differentiation and/or infiltration (53 to 72%) with lower GATA3 expression (16 to 26%) in both the lung and BAL (SI Appendix, Fig. S4 G–P). As with the HDM challenge model, the impact of Gata3 +674/762 deletion on Th2 cells was markedly less pronounced than on ILC2s in both the lung and BAL following S. venezuelensis infection. These results, and the persistence of some residual ILC2s, may explain why the time course of helminth clearance was similar in Gata3 +674/762Δ/Δ mice compared to WT littermates (SI Appendix, Fig. S4Q). In sum, the type 2 inflammatory response following exposure to HDM extract and S. venezuelensis infection is defective in Gata3 +674/762Δ/Δ mice, and the defect is more pronounced in ILC2s compared with Th2 cells.
Lastly, we used a third model of type 2 pulmonary inflammation, based on antigen adjuvanted allergen challenge via intraperitoneal (i.p.) systemic ovalbumin (OVA)–alum administration followed by i.n. challenge with OVA (Fig. 3I). The rationale for this additional model was that, unlike HDM and S. venezuelensis, OVA–alum-induced allergic airway inflammation was previously suggested to be largely independent of ILC2s (29, 46). Strikingly, in contrast with HDM and S. venezuelensis–challenged mice, the extent of eosinophilia in the lung and BAL was comparable between WT littermate and Gata3 +674/762Δ/Δ mice treated with the OVA–alum protocol (Fig. 3J). Furthermore, despite the persistent reduction in cell numbers (∼75%) and GATA3 expression (∼45%) by ILC2s, Th2 cells showed unaltered expansion, differentiation, infiltration, GATA3 expression, and type 2 cytokine production in the lung and BAL of OVA–alum-treated Gata3 +674/762Δ/Δ mice compared with WT littermate mice (Fig. 3 K–N). These results not only support prior observations that ILC2s are largely dispensable in the systemic OVA–alum model, but also demonstrate that Gata3 +674/762Δ/Δ Th2 cells are fully capable of eliciting a normal, robust type 2 inflammatory response.
Gata3 +674/762Δ/Δ Mice Exhibit a Profound Cell-Intrinsic ILC2 Defect and a Modest, Partially Cell-Extrinsic Th2 Cell Defect.
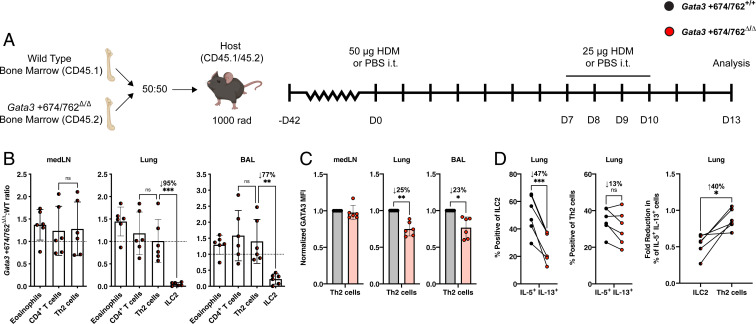
In Gata3 +674/762Δ/Δ mice, extrinsic factors could contribute to the impaired differentiation and response of Th2 cells to HDM challenge and S. venezuelensis infection. Though CD4+ T cell numbers were normal and did not require Gata3 +674/762 at homeostasis or in competition with WT cells (Figs. 1I and 2F), the deficiency in ILC2 numbers and early type 2 cytokine production in Gata3 +674/762Δ/Δ mice may hinder the proper priming and progression of an adaptive type 2 response (29–31, 46). To address this possibility, we evaluated the intrinsic differentiation capacity of Gata3 +674/762Δ/Δ Th2 cells through a competitive (Gata3 +674/762Δ/Δ:WT) mixed bone marrow chimera model where mice were sensitized and challenged i.t. with HDM extract (Fig. 4A). In this model, the differentiation of Gata3 +674/762Δ/Δ Th2 cells occurs in the presence of a WT ILC2 compartment, permitting an assessment of the intrinsic type 2 differentiation capability of WT and Gata3 +674/762Δ/Δ CD4+ T cells. As expected from unchallenged mixed bone marrow chimeras, the reconstitution frequency of Gata3 +674/762Δ/Δ and WT eosinophils and CD4+ T cells was equivalent while ILC2s from Gata3 +674/762Δ/Δ BM were profoundly outcompeted 1:20 in the lung and 1:4 in the BAL by WT ILC2s (Fig. 4B). In contrast to ILC2s, however, differentiated Th2 cells of both genotypes were equally represented in the mediastinal lymph node (medLN), lung, and BAL. Nevertheless, Gata3 +674/762Δ/Δ Th2 cells expressed ∼25% lower levels of GATA3 in the lung and BAL (Fig. 4C). Furthermore, while Gata3 +674/762Δ/Δ ILC2s were intrinsically and profoundly deficient in their ability to produce both IL-5 and IL-13 (47% reduction compared to WT ILC2s), Th2 cells exhibited only a minor nonsignificant defect (13% reduction compared to WT Th2 cells) (Fig. 4D). When the deficiency in ILC2 and Th2 cell cytokine production was directly evaluated in a pairwise comparison, Gata3 +674/762Δ/Δ ILC2s were found to be significantly more impaired than Gata3 +674/762Δ/Δ Th2 cells (40% lower than Th2 cells). Taken together, these findings support the conclusion of a predominant and profound cell-intrinsic impact of Gata3 +674/762 deletion on ILC2 function and a modest, largely cell-extrinsic impact on Th2 cells that may be secondary to ILC2 dysfunction. This interpretation is also consistent with the preserved Th2 cell frequency and function in the ILC2-independent model of OVA–alum allergic inflammation.
Fig. 4.
Cell-intrinsic impact of Gata3 +674/762 deletion on Th2 cell differentiation and function. (A) Schematic for HDM-induced allergic airway inflammation in congenically marked mixed bone marrow chimeras. (B) Gata3 +674/762Δ/Δ:WT reconstitution ratio for the indicated populations in the medLN, lung, and BAL from HDM-treated mixed bone marrow chimeric mice. (C) Summary plots of GATA3 MFI in medLN, lung, and BAL Th2 cells. (D) Summary data for frequency of IL-5/IL-13 cytokine double producers from lung ILC2s and Th2 cells, and summary data showing the pairwise comparison of the fold reduction in the frequency of IL-5/IL-13 cytokine double producers for lung ILC2s (Gata3 +674/762Δ/Δ:WT) vs. lung Th2 cells (Gata3 +674/762Δ/Δ:WT) in mixed bone marrow chimeras. Dots represent individual mice; n = 6 for mixed bone marrow chimeras and data are pooled from multiple independent experiments; data are presented as mean ± SEM. Values of 0 were converted to a value of 1 on a log scale. Statistical comparison was performed via paired t test or one-way ANOVA. *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant.
Characterization of Gata3 +761/762, a Regulatory Element Recapitulating Gata3 Regulation in ILC2s.
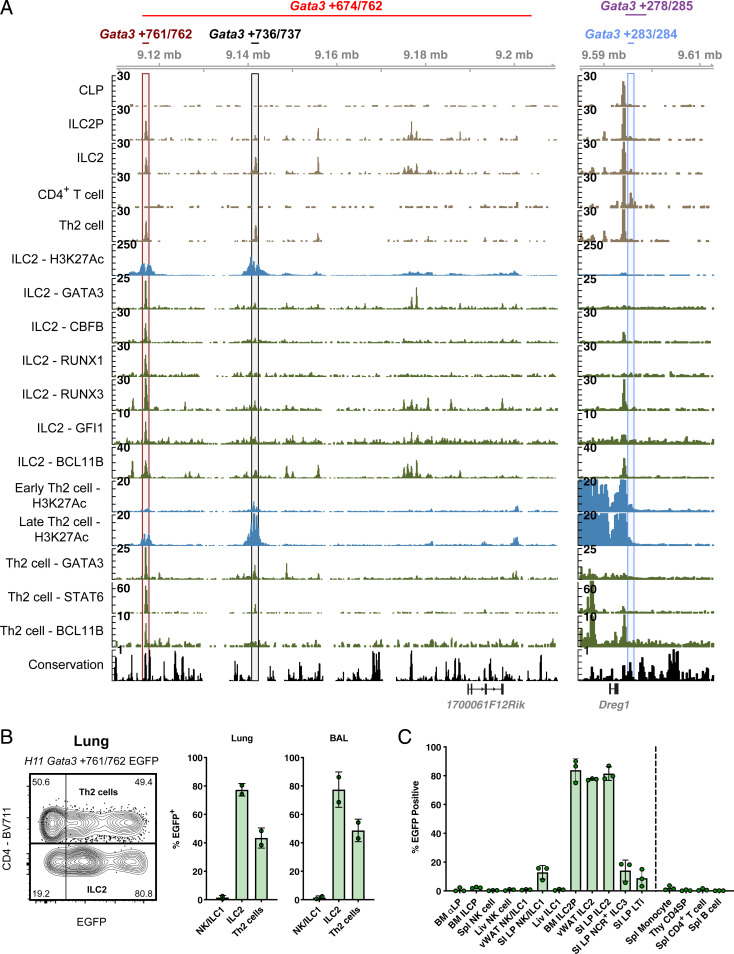
Our results indicated that the Gata3 +674/762 region controlled the proper differentiation and function of ILC2s and, to a minor degree, Th2 cells. To identify which elements within Gata3 +674/762 contributed to the regulatory function, we utilized public chromatin immunoprecipitation (ChIP)-seq data to assess additional parameters typical of enhancer elements. Alignment of histone 3 K27 acetylation (H3K27Ac) and TF binding in ILC2s and Th2 cells, in addition to genomic conservation, revealed a top candidate subdomain, Gata3 +761/762, with enhancer-like characteristics in ILC2s and Th2 cells (Fig. 5A, maroon window). Gata3 +761/762 was conserved in placental mammalian genomes and its flanking regions accumulated H3K27Ac marks (54, 55). Furthermore, this segment was bound by several key type 2 lineage-determining TFs, including GATA3, BCL11B, GFI1, and STAT6, as well as other TFs such as RUNX1, RUNX3, and CBFB (15, 54–58). Another putative enhancer, Gata3 +736/737, exhibited somewhat similar though markedly weaker features and did not show genomic conservation (Fig. 5A, black window). Notably, neither H3K27Ac nor any of the analyzed TFs accumulated at the 1.2-kb core element of Gata3 +278/285, Gata3 +283/284, in mature ILC2s. or differentiated Th2 cells, consistent with the distinct functions of these enhancer regions (Fig. 5A, cornflower blue window). We next scanned for immune-specific TF motifs present within the Gata3 +761/762, Gata3 +736/737, and Gata3 +283/284 regions using the TRANSFAC database to gain insight into potential regulatory mechanisms (SI Appendix, Fig. S5A) (59). Strikingly, Gata3 +761/762 contained a multitude of predicted GATA3 motifs, seven in total, while Gata3 +736/737 and Gata3 +283/284 only contained three and one GATA3 motif, respectively. Enrichment for GATA3 motifs within Gata3 +761/762 agrees with the observed ChIP-seq enrichment seen in ILC2s and Th2 cells (Fig. 5A), suggesting a potential autoregulatory role for GATA3 in type 2–specific Gata3 expression (41, 60, 61). In sum, these results indicate that Gata3 +674/762, and Gata3 +761/762 in particular, are enriched for TF binding in vivo and binding motifs in silico, suggestive of type 2–specific function.
Fig. 5.
Characterization of Gata3 +674/762, and a GATA3 binding element in Gata3 +674/762. (A) ATAC-seq accessibility coverage (beige), ChIP-seq histone modification (blue), ChIP-seq transcription factor binding (green), and UCSC conservation (black) tracks for the Gata3 +674/762 and Gata3 +278/285 regions in the indicated populations from public sequencing data (2, 15, 54–58). Maroon, black, and cornflower blue windows represent Gata3 +761/762, Gata3 +736/737, and Gata3 +283/284 regions, respectively. (B) Representative flow cytometry plot of H11 Gata3 +761/762 EGFP reporter expression in ILC2s (pregated on CD45.2+CD19−CD11c−CD3ε−TCRβ−IL-7Rα+CD90.2+CD25+IL-33Rα+) and Th2 cells (pregated on CD45.2+CD3ε+TCRβ+CD90.2+CD4+IL-33Rα+) from the lung following HDM challenge. Summary bar graph shows H11 Gata3 +761/762 EGFP reporter expression in ILC2s, Th2 cells, and NK/ILC1 from the lung and BAL. (C) H11 Gata3 +761/762 EGFP reporter expression in various lymphocyte populations from different tissues as indicated. Dots represent individual mice; n ranging from 2 to 3 in one experiment from independent founder (F0) mice; data are presented as mean ± SEM.
To further dissect the Gata3 +674/762 enhancer region, we next generated deletion strains with CRISPR/Cas9 targeting (SI Appendix, Fig. S5B). However, deletion of Gata3 +674/710 or the complementary Gata3 +710/762 segment appeared to only marginally, or nonsignificantly, compromise ILC2 numbers in the lung, failing to recapitulate the magnitude of the ILC2 defect associated with the larger Gata3 +674/762 deletion (SI Appendix, Fig. S5 C and D), indicating the presence of redundant regulatory elements, a well-established feature of many important enhancers (62). Moreover, these results demonstrated that while lncRNA 1700061F12Rik was expressed in ILC2s (SI Appendix, Fig. S1A), it was not necessary for ILC2 development. To determine the sufficiency of Gata3 +761/762, the top candidate regulatory element within Gata3 +674/762, to contribute to the regions characterized type 2–specific Gata3 regulatory function, we generated in vivo enhancer reporter mice via CRISPR/Cas9-mediated integration (18). Genomic integration of an enhancer element upstream of a minimal promoter, such as the heat shock protein 68 (HSP68) minimal promoter, has been used to track in vivo enhancer activity by driving the expression of Cre, lacZ, fluorescent proteins, or coding genes (63–66). However, traditional methods used to generate transgenic mice are hampered by concerns of uncontrolled integration and multiple copy number. Site-specific integration into a safe harbor locus, such as Rosa26 or H11, provides a means to control these factors while additionally increasing efficiency and minimizing time to generation (63, 67). We therefore applied a CRISPR/Cas9-mediated integration strategy to target the H11 locus (SI Appendix, Fig. S5E) (18). To this end, we generated H11 Gata3 +761/762 enhanced green fluorescent protein (EGFP) reporter mice and characterized the pattern of EGFP expression in ILCs and T cells. Following treatment of H11 Gata3 +761/762 EGFP reporter mice with HDM extract as described before (Fig. 3A), we found that ILC2s and, to a lesser degree Th2 cells, in the lung and BAL were marked by EGFP expression, whereas group 1 ILCs were not (Fig. 5B). More broadly, we observed high levels of EGFP expression specifically in ILC2s from the BM, SI LP, and vWAT, but not in group 1 or 3 ILCs, thymocytes, CD4+ T cells, or B cells (Fig. 5C). From these observations, we concluded that the Gata3 +761/762 element is sufficient to recapitulate the pattern of type 2–specific Gata3 enhancer activity in ILC2s and Th2 cells.
Discussion
ILC2s and Th2 cells express a similar GATA3-driven type 2 helper effector program and contribute to a variety of allergic and helminthic inflammatory processes, but their differentiation occurs in very different contexts (2, 3). While optimal Th2 cell differentiation from naïve CD4+ T cells occurs in the lymph nodes following antigen exposure and IL-4–mediated activation of STAT6 (42, 61, 68, 69), ILC2s differentiate from precursors in the BM and peripheral tissues (16, 70) and are maintained in the periphery at homeostasis independently of STAT6 or exposure to type 2 cytokines (71). To understand the mechanisms underlying these differences, we leveraged ATAC-seq chromatin accessibility data in combination with CRISPR/Cas9-mediated deletion to identify cis-regulatory elements controlling the Gata3 locus. Our studies identified a distal regulatory region, Gata3 +674/762, that primarily controls the frequency and differentiation of ILC2s and, to a lesser and mostly indirect degree, Th2 cells. Chromatin accessibility within Gata3 +674/762 coincided with elevated Gata3 expression in ILC2s and Th2 cells (2, 10, 18, 20, 21), and deletion of this region specifically and profoundly impacted the frequency and function of ILC2s, and to a more minor extent Th2 cells, leaving group 1 and 3 ILCs as well as CD4+ T cells unperturbed. In comparison, the previously identified enhancer Gata3 +278/285 did not impact ILC2 differentiation or function, but instead contributed broadly to the development of all ILC subsets. Following HDM allergen, S. venezuelensis helminth, or OVA–alum challenge, Gata3 +674/762Δ/Δ mice were variably compromised in their ability to mount a type 2 inflammatory response. Notably, while ILC2s were profoundly and intrinsically impaired by deletion of Gata3 +674/762 in all contexts, the impact of enhancer deletion on Th2 cells was milder or completely absent, depending on the nature of the type 2 challenge. The limited Th2 cell defect stemmed from both an extrinsic reduction in ILC2 numbers and function and a partial cell-intrinsic deficiency. The lack of a substantial Th2 cell defect explained the generally modest impact of Gata3 +674/762 deletion on eosinophilia and goblet cell hyperplasia that characterize the overall type 2 inflammatory response. Lastly, we identified a highly conserved element of the Gata3 +674/762 region, Gata3 +761/762, that was sufficient to recapitulate key features of the entire region, including its specific activity in ILC2s, upon incorporation into an in vivo enhancer reporter system.
Functionally, Gata3 +674/762 controlled the number, type 2 differentiation, and function of ILC2s at homeostasis and during a variety of type 2 inflammatory challenges, including HDM, S. venezuelensis, and OVA–alum. Lung ILC2s were profoundly decreased by 75 to 96% at homeostasis and after allergic and helminthic challenges in mice bearing a deletion of Gata3 +674/762. Furthermore, the few residual ILC2s in Gata3 +674/762Δ/Δ mice were impaired by ∼50% in GATA3 expression, a level previously shown to significantly alter their ability to produce type 2 cytokines (17), consistent with a 64% reduction in IL-5/IL-13 double-producer lung ILC2s. In contrast, Th2 cells showed a more limited and mostly indirect dependency on Gata3 +674/762 for expansion and differentiation following allergic and helminthic challenge. Whereas lung ILC2s were profoundly decreased at homeostasis and after type 2 inflammatory challenges in mice bearing a deletion of Gata3 +674/762, Th2 cells were diminished by 53 to 86% following HDM or S. venezuelensis challenge, or not decreased at all in the case of the ILC2-independent OVA–alum challenge model (29, 46). Furthermore, this decrease in Th2 cells was largely corrected in a mixed bone marrow chimera setting, indicating a predominantly cell-extrinsic origin for the Th2 cell defect in Gata3 +674/762Δ/Δ mice that may be secondary to the deficiency in type 2 cytokine production from ILC2s. For example, production of IL-13 by ILC2s has been suggested to promote dendritic cell migration to the draining lymph node (30, 72). Separately, production of IL-4 by Th2 cells could further enhance Th2 cell differentiation in an autocrine or paracrine manner (61, 73). Accordingly, our findings indicated that deletion of Gata3 +674/762 induced a major numerical and functional defect in ILC2s but had only a partial, mostly indirect impact on Th2 cells, explaining the relatively minor overall functional impact of the Gata3 +674/762 deletion on a variety of type 2 responses, particularly those, like OVA–alum, that do not appear to involve significant ILC2 contribution (29, 46). Thus, our study provides a genetic basis for the distinct mechanisms involved in driving innate vs. adaptive type 2 responses, with potential evolutionary implications for their association with distinct types of immune challenges.
Our study also established that the previously described Gata3 +278/285 enhancer, termed TCE7.1 (21, 28), had a very distinct pattern of dynamic accessibility and function compared with Gata3 +674/762. Gata3 +278/285 appeared to control the low/medium levels of GATA3 expression required for the development of early ILC precursors and thymocytes but had little impact on the acquisition of the type 2 helper effector program in ILC2s, including the elevated GATA3, IL-33Rα, and cytokine expressions that are driven instead by Gata3 +674/762 in ILC2s.
Our efforts to further dissect the active elements in the Gata3 +674/762 enhancer region were partially hampered by the finding that distinct portions of this region seemed to contribute to enhancer function in an apparently redundant manner (SI Appendix, Fig. S5 C and D). Deletion of either Gata3 +674/710 or the complementary Gata3 +710/762 segments did not significantly impair or only marginally impaired enhancer function as judged by lung ILC2 numbers remaining within twofold of WT littermate ILC2s. The presence of separate but redundant regulatory elements is a well-characterized phenomenon across both vertebrates and invertebrates that confers robustness, patterning precision, and evolvability to gene expression (62). Nevertheless, it was possible to characterize at least one potent regulatory element, Gata3 +761/762, contained within Gata3 +674/762 based on several key properties. First Gata3 +761/762 displayed chromatin accessibility in ILC2s and exhibited markers of activation such as H3K27 acetylation (2, 54). Second, binding of GATA3, CBFB, RUNX1 and RUNX3, BCL11B, and GFI1 was enriched within Gata3 +761/762, as determined by published ILC2 ChIP-seq data (15, 55–57). Lastly, Gata3 +761/762 showed high sequence conservation in placental mammals. Based on these suggestive features, we established an in vivo enhancer reporter that demonstrated specific and high levels of reporter expression in ILC2s, and to a lesser degree, Th2 cells, but not in various other innate or adaptive cell subsets. Notably, this enhancer reporter system may be exploited, for example, to drive the expression of an inducible Cre or diphtheria toxin receptor, providing “next-generation” tools to assess or manipulate the distinct contribution of ILC2s toward type 2 inflammatory challenges in the context of an unperturbed adaptive immune response.
Thus, while further dissection of the distal Gata3 +674/762 region is warranted, our studies demonstrate and partially characterize the presence of a Gata3 enhancer controlling ILC2 development and function that is largely dispensable for Th2 cells and suggest the existence of additional, distinct regulatory elements controlling Gata3 expression in Th2 cells. Furthermore, in addition to the GATA3 binding site identified by direct ChIP-seq analysis, motif analysis in accessible portions of this region revealed several additional putative GATA3 binding motifs. These observations support the possibility that GATA3 may exert a prominent positive feedback on its own expression (41, 60, 61), providing a potential mechanistic explanation for achieving the elevated levels of GATA3 required to support the type 2 helper effector program of ILC2s.
Materials and Methods
Mice.
C57BL/6J (CD45.2+; stock No. 000664) and B6.SJL-Ptprca Pepcb/Boy (CD45.1+; stock No. 002014) mice were purchased from The Jackson Laboratory and maintained in house. Male and female littermate mice ranging from 6 to 8 wk were used in all experiments of this study. All mice were bred and housed in a specific pathogen-free facility at the University of Chicago. Experiments were conducted under the guidelines of the University of Chicago Institutional Animal Care and Use Committee.
Generation of Enhancer Deletion and Reporter Mice.
Gata3 +674/762Δ/Δ, Gata3 +674/710Δ/Δ, Gata3 +710/762Δ/Δ, and Gata3 +278/285Δ/Δ mice were generated via CRISPR/Cas9-mediated deletion. Guide sites were selected using IDT design tools, and Alt-R CRISPR/Cas9 crRNA, tracrRNA, and Cas9 nuclease were purchased from IDT. Targeting strategies and crRNA sequences are displayed in SI Appendix, Figs. S1C, S3A, and S5B. For microinjections, guide RNA (gRNA) was assembled with crRNA and tracrRNA according to the manufacturer’s instructions and added to the final injection mix at 50 ng/μL and 50 ng/μL Cas9 per guide in embryo grade water. Mixes were injected into the nuclei of C57BL/6J embryos. Successful deletions were determined by PCR and sequence verified. Deletion mice were maintained on the C57BL/6J background and backcrossed for four generations unless otherwise indicated. H11 enhancer reporter mice were generated via CRISPR/Cas9-mediated insertion as before (SI Appendix, Fig. S5E) (18). Targeting strategy and crRNA sequence are displayed in SI Appendix, Fig. S5E. Briefly, the Gata3 +761/762 sequence was cloned upstream of the murine minHSP68 promoter and EGFP, which was then inserted into a pUC19 vector between 1-kb and 3-kb asymmetric homology arms. Microinjection mixes were assembled as above with 20 ng/μL gRNA, 50 ng/μL Cas9 nuclease, and 12.5 ng/μL plasmid in embryo grade water. Successful integrations were determined by targeting PCR using two primer sets that generate a product spanning the insert out beyond the homology arms, which was sequence verified.
Preparation of Cell Suspensions.
Bone marrow was collected from the femur, tibia, and ilium (six bones in total), crushed with the syringe plunger flange, passed through a 70-μm cell strainer, and resuspended in fluorescence-activated cell sorting (FACS) buffer (1× Hank's Balanced Salt Solution [HBSS] [Gibco], 0.25% bovine serum albumin [BSA] [Millipore-Sigma], 50 ng/mL DNase, and 5 mM sodium azide [Millipore-Sigma]). Spleens, thymi, and lymph nodes were prepared by passing through a 70-μm cell strainer and resuspending in FACS buffer. Adult livers were passed through a 70-μm cell strainer and resuspended in FACS buffer. Fat was removed by centrifugation in 0.45% Percoll (Millipore-Sigma). Red blood cells (RBC) were removed with 1× RBC lysis buffer (Thermo Fisher). The lung was perfused with 1× phosphate buffered saline (PBS) (Corning), chopped into small pieces, and digested by shaking (250 rpm) at 37 °C for 20 min in Roswell Park Memorial Institute (RPMI) medium (HyClone) with 650 U/mL Collagenase A (Roche) and 0.01% DNase (Millipore-Sigma). The SI LP was prepared by removing Peyer’s patches, washing out intestinal contents with ice cold 1× PBS, and cutting length and width wise into small pieces. Intraepithelial lymphocytes were removed by two 37 °C 15 min shakes (250 rpm) in RPMI with 5 mM ethylenediaminetetraacetic acid (EDTA) (Fisher Scientific) and 1% fetal bovine serum (FBS) (Atlanta Biologicals), and lamina propria was digested by shaking (250 rpm) at 37 °C for 30 min in RPMI with 20% FBS, 0.5 mg/mL collagenase A, and 0.17 mg/mL DNase. vWAT was digested by shaking (250 rpm) at 37 °C for 45 min in RPMI with 1 mg/mL collagenase A. Digests for the lung, SI LP, and vWAT were passed through 70-μm or 100-μm cell strainers, followed by excess fat removal and RBC lysis as above. BAL was prepared by inserting a cannula into the trachea followed by flushing of the lungs four times with 1× PBS 5 mM EDTA to collect a total of 3.5 mL BAL fluid.
Flow Cytometry.
Single cell suspensions were incubated with BD FcBlock for 15 min on ice. The following fluorochrome- or biotin-conjugated antibodies were used: α4β7 (DATK32), CCR6 (29-2L17), CD11b (M1/70), CD11c (N418), CD45.1 (A20), CD45.2 (104), CD90.2 (Thy1.2; 53-2.1), CD127 (IL-7Rα; A7R34), GR-1 (RB6-8C5), ICOS (C398.4A), IL-4 (11B11), IL-17RB (9B10), IL-33Rα (DIH9), KLRG1 (MAFA), NK1.1 (PK136), NKp46 (29A1.4), PD-1 (29F.1A12), TCRγδ (GL3), mouse IgG1 κ-chain (MG1-45), mouse IgG2a κ-chain (MG2a-53), rat IgG1 κ-chain (RTK20-71), rat IgG2a κ-chain (RTK27-58), and rat IgG2b κ-chain (RTK45-30) (all from BioLegend); B220 (RA3-6B2), CD3ε (145-2C11), CD4 (GK1.5), CD8α (53-6.7), CD11b (M1/70), CD11c (HL3), CD19 (1D3), CD25 (PC61), CD45.2 (104), CD49a (Ha31/8), CD117 (cKit, 2B8), CD121a (35F5), Flt3 (A2F10.1), GR-1 (RB6-8C5), IL-5 (TRFK5), Ly6A/E (Sca-1, D7), NK1.1 (PK136), Siglec-F (E50-2440), TCRβ (H57-597), and Ter119 (TER-119) (all from BD Biosciences); GATA3 (TWAJ), IL-13 (eBio13A), and FOXP3 (FJK-16s) (all from Thermo Fisher); and NRP-1 (761705) from R&D Systems. To exclude dead cells, Zombie NIR, yellow, or violet fixable viability dye (BioLegend) was added to live cells. Intracellular transcription factor staining was performed using the Foxp3 Transcription Factor Staining Buffer Kit (Thermo Fisher) according to the manufacturer’s instructions. Staining for cytokines was performed using the BD Cytofix/Cytoperm Fixation/Permeabilization Kit according to the manufacturer’s instructions with fixation in 4% paraformaldehyde (PFA) (Electron Microscopy Science). Cells were blocked with unlabeled isotype-matched antibody (above) and negative controls were prepared using 20-fold excess unlabeled antibody to the transcription factor or cytokine (cold competitor). Samples were acquired on a 4-laser LSRII (BD Bioscences) and analyzed using FlowJo software (TreeStar). The following lineage (Lin) stain was used for the BM: B220, CD3ε, CD4, CD8α, CD11b, CD11c, CD19, GR-1, NK1.1, TCRβ, and Ter119. Unless otherwise indicated, cell populations were identified as follows while excluding alternate lineages: BM CLP (Lin−IL-7Rα+Flt3+Sca-1+cKit+), BM αLP (Lin−α4β7+CD90.2−IL-7Rα+Tcf7mCherry-PD-1−Flt3+), BM rEILP (Lin−α4β7+CD90.2−IL-7Rα−Tcf7mCherry+PD-1−), BM iILCP (Lin−α4β7+CD90.2−IL-7Rα−Tcf7mCherry+PD-1+), BM ILCP (Lin−α4β7+IL-7Rα+Tcf7mCherry+PD-1+), BM ILC2P (Lin−α4β7+IL-7Rα+CD90.2+ICOS+PD-1−), splenic NK cells (CD45.2+CD19−CD3ε−TCRβ−NK1.1+), liver NK cells (CD45.2+CD19−CD3ε−TCRβ−NK1.1+DX5+CD49a−), liver ILC1s (CD45.2+CD19−CD3ε−TCRβ−NK1.1+DX5−CD49a+), lung and vWAT NK/ILC1 (CD45.2+CD19−CD11c−CD3ε−TCRβ−NK1.1+), lung and BAL ILC2s (CD45.2+CD19−CD11c−CD3ε−TCRβ−IL-7Rα+CD90.2+CD25+IL-33Rα+), vWAT ILC2s (CD45.2+CD19−CD11c−CD3ε−TCRβ−IL-7Rα+CD90.2+CD25+IL-33Rα+KLRG1+), SI LP ILC2s (CD45.2+CD19−CD11c−CD3ε−TCRβ−IL-7Rα+CD90.2+KLRG1+Sca-1+IL-17RB+), SI LP NCR+ ILC3s (CD45.2+CD19−CD11c−CD3ε−TCRβ−IL-7Rα+CD90.2+CD121a+CCR6−NKp46+), SI LP LTis (CD45.2+CD19−CD11c−CD3ε−TCRβ−IL-7Rα+CD90.2+CD121a+CCR6+NKp46−NRP1+), SI LP ILC3s (CD45.2+CD19−CD11c−CD3ε−TCRβ−IL-7Rα+CD90.2+CD121a+), Thy ETP (B220−CD11b−CD11c−CD19−GR-1−CD4−CD8α−TCRγδ−CD1d-Tet−TCRβ−CD25−CD44+), Thy CD4SP (B220−CD11b−CD11c−CD19−GR-1−CD4+CD8α−TCRγδ−CD1d-Tet−), Thy CD8SP (B220−CD11b−CD11c−CD19−GR-1−CD4−CD8α+TCRγδ−CD1d-Tet−TCRβ+), splenic T cells (CD45.2+CD3ε+TCRβ+, CD4+ or CD8α+), lung and BAL Th2 cells (CD45.2+CD3ε+TCRβ+CD90.2+CD4+IL-33Rα+GATA3+), medLN Th2 cells (CD45.2+CD3ε+TCRβ+CD90.2+CD4+GATA3+), splenic B cells (CD45.2+B220+CD19+), splenic neutrophils (CD45.2+CD11b+GR-1+FSCHiSSCHi), lung, vWAT, BAL, and medLN eosinophils (CD45.2+Siglec-F+SSCHi), lung and BAL AMac (CD45.2+CD11c+Siglec-F+), vWAT GATA3+ Treg cells (CD45.2+CD3ε+TCRβ+CD90.2+CD4+FOXP3+GATA3+IL-33Rα+KLRG1+), and vWAT GATA3− Treg cells (CD45.2+CD3ε+TCRβ+CD90.2+CD4+FOXP3+GATA3−).
Mixed Bone Marrow Chimeras.
To generate mixed chimeras, bone marrow was isolated as described above for Gata3 +674/762Δ/Δ (CD45.2) mice and WT (CD45.1 or CD45.1/0.2) mice, depleted of T cells, and mixed at a 50:50 ratio. Ten million mixed cells were injected retroorbitally into lethally irradiated (1,000 rad) CD45.1/0.2 or CD45.1 recipients to allow for discrimination of donor Gata3 +674/762Δ/Δ and WT cells from host cells. Host mice were allowed to recover for 6 wk prior to analysis or treatment. Populations were normalized to the reconstitution frequency of splenic neutrophils or splenic B cells.
IL-33 Challenge.
A total of 500 ng of IL-33 (BioLegend) in PBS was administered i.n. in 40 μL to isoflurane (Henry Schein) anesthetized mice on days 0, 1, and 2, followed by analysis on day 3 (SI Appendix, Fig. S1L). Control mice were administered PBS alone.
HDM Challenge.
HDM extracts (Stallergenes Greer Lenoir) were suspended in 1× sterile endotoxin-free PBS (Sigma) to a concentration of 4 mg/mL. Lots were titrated to induce a minimum of 2 × 106 eosinophils in the BAL of C57BL/6 mice on day 13 using an i.t. challenge model (Figs. 3A and 4A). Briefly, mice were anesthetized with i.p. injection of ketamine/xylazine (Covetrus) and sensitized via administration of 50 μg HDM extract in 50 μL 1× sterile endotoxin-free PBS through i.t. instillation on day 0. After 7 d, mice were challenged i.t. with 25 μg HDM extract in 50 μL 1× sterile endotoxin-free PBS on days 7 through 10 followed by analysis on day 13. Control mice were treated with PBS alone during the sensitization and challenge phases.
S. venezuelensis Passage and Infection.
S. venezuelensis was propagated in NOD-scid IL2Rγ-/- (NSG) mice (The Jackson Laboratory) by s.c. infection with 10,000 larvae. Prior to infection of mice, feces were collected and spread on Whatman paper, and the paper was placed into a beaker with 28 °C water. Hatching live larvae were collected after 4 d. Mice were s.c. infected with 700 L3 larvae per mouse. For egg count, feces were collected, weighed, and homogenized with counts normalized to feces weight.
OVA–Alum Challenge.
Mice were treated using a 23-d model of sensitization and challenge as previously described (Fig. 3I) (46). Briefly, 50 μL of 1 mg/mL grade V OVA (Sigma) in 1× PBS was mixed with 50 μL of Imject alum (Thermo Fisher), and 100 μL of the mixture was injected i.p. on days 0, 7, and 14 for sensitization. Subsequently, mice were challenged i.n. with 100 μg OVA in 40 μL of 1× PBS or PBS only on days 20 to 22 and analyzed on day 23.
Histology.
Subsequent to perfusion, as described above, the left lobe of the lung was removed and fixed in 10% neutral buffered formalin (Azer Scientific) for a minimum of 24 h before transfer to 70% ethanol (Fisher Scientific) for storage. Lobes were embedded in paraffin and cut into 5-μm sections before staining with periodic acid–Schiff (PAS). Histology slides were scanned using the Pannoramic SCAN 40× Whole Slide Scanner and analyzed with QuPath whole slide image analysis software (https://qupath.github.io/). Histology slides were blinded for PAS scoring and scored on a scale from 0 to 5 as follows: 0 = no PAS staining, 1 = 0 to 25% PAS+ cells, 2 = 26 to 50% PAS+ cells, 3 = 51 to 75% PAS+ cells, 4 = 76 to 100% PAS+ cells, and 5 = 100% PAS+ cells and evidence of mucus plugs.
ATAC-Seq Sample Preparation.
ATAC-seq was performed as described in the published Omni-ATAC protocol (74). Briefly, 5,000 to 10,000 cells were sorted into FACS buffer and washed twice with 1 mL ice-cold ATAC-seq resuspension buffer (RSB) (10 mM Tris, pH 7.4 [Invitrogen], 10 mM NaCl [Sigma], 3 mM MgCl2 [Sigma]) by centrifugation at 0.5 × g for 5 min at 4 °C. After centrifugation, the supernatant was carefully removed to avoid the cell pellet, which was then gently resuspended in 50 μL ATAC-seq RSB containing 0.1% Nonidet P-40 (Roche), 0.1% Tween-20 (Sigma), and 0.1% digitonin (Sigma). The lysis reaction was incubated on ice for 3 min, 1 mL ATAC-seq RSB containing 0.1% Tween-20 was added, and the nuclei were centrifuged at 0.5 × g for 10 min at 4 °C. The supernatant was subsequently removed, and the pelleted nuclei were gently resuspended in 50 μL transposition mix (25 μL 2× TD buffer [Illumina], 2.5 μL Tn5 transposase [Illumina], 16.5 μL 1× PBS, 0.5 μL 1% digitonin, 0.5 μL 1% Tween-20, and 5 μL water). Transposition reactions were incubated for 30 min in a 37 °C water bath. Zymo DNA Clean and Concentrator kits were used to clean up the transposition reactions, and libraries were prepared as previously described (75). Libraries were sequenced by 50-bp single-end, dual-index sequencing on an Illumina HiSEq. 4000.
Bioinformatic Analysis.
Publicly available data were downloaded from the National Center for Biotechnology Information Sequence Read Archive database (2, 15, 54–58). Reads were aligned to the mouse genome (mm10) with Bowtie and duplicate reads were removed using Picard Tools. Bedgraphs were generated using HOMER (76) and files were converted to bigWig format for visualization using the University of California Santa Cruz (UCSC) bedGraphToBigWig program. The R package Gviz was used for coverage track visualization.
Statistical Analysis.
Student’s t test, multiple t tests controlling for false discovery rate, and one-way ANOVA with Tukey’s multiple comparisons test were performed in GraphPad Prism 8.
Online Supplemental Material.
SI Appendix, Fig. S1 provides a schematic for the generation of Gata3 +674/762Δ/Δ mice and analysis of select immune cell populations (related to Figs. 1 and 2). SI Appendix, Fig. S2 provides ATAC-seq coverage tracks across the Gata3 locus for select immune cell populations (related to Fig. 1). SI Appendix, Fig. S3 provides a schematic for the generation of Gata3 +278/285Δ/Δ mice and analysis of select immune cell populations. SI Appendix, Fig. S4 provides additional phenotyping of immune cell populations from HDM-challenged mice and S. venezuelensis–infected mice (related to Fig. 3). SI Appendix, Fig. S5 provides in silico transcription factor motif data, a schematic for the generation of Gata3 +674/710Δ/Δ and Gata3 +710/762Δ/Δ mice, analysis of lung ILC2s in said mice, and a schematic for generating H11 enhancer reporter mice (related to Fig. 5).
Supplementary Material
Acknowledgments
We thank D. Esterhazy (University of Chicago) for providing S. venezuelensis for infection experiments; A. Hoffman (University of Chicago) for technical guidance with vWAT preparation; and C. Stamper (University of Chicago) for technical guidance with i.n. treatment. We are grateful to L. Degenstein (University of Chicago) and the University of Chicago Transgenics Core for support with microinjections; the University of Chicago DNA Sequencing and Genotyping Core for sequencing of reporter plasmid constructs; the University of Chicago Genomics Core for sequencing of ATAC-seq samples; the University of Chicago Human Tissue Resource Center for processing, embedding, and staining of histology samples; and the University of Chicago Integrated Microscopy Core for whole slide scanning. Schematics in Figs. 2E and 5A depicting a mouse and mouse femurs were created with images from http://BioRender.com. This work was supported by the NIH (Grants R37 AI127518, R01 AI144094, and U01 AI125250) and support funds from the University of Chicago Dean’s office.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2106311118/-/DCSupplemental.
Data Availability
ATAC-seq data have been deposited in Gene Expression Omnibus (GSE169542) (77). All other study data are included in the article and/or supporting information.
References
- 1.Vivier E., et al., Innate lymphoid cells: 10 years on. Cell 174, 1054–1066 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Shih H.-Y., et al., Developmental acquisition of regulomes underlies innate lymphoid cell functionality. Cell 165, 1120–1133 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Dyken S. J., et al., A tissue checkpoint regulates type 2 immunity. Nat. Immunol. 17, 1381–1387 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wan Y. Y., GATA3: A master of many trades in immune regulation. Trends Immunol. 35, 233–242 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu J., GATA3 regulates the development and functions of innate lymphoid cell subsets at multiple stages. Front. Immunol. 8, 1–8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hosoya T., et al., GATA-3 is required for early T lineage progenitor development. J. Exp. Med. 206, 2987–3000 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ting C.-N., Olson M. C., Barton K. P., Leiden J. M., Transcription factor GATA-3 is required for development of the T-cell lineage. Nature 384, 474–478 (1996). [DOI] [PubMed] [Google Scholar]
- 8.Pai S.-Y., et al., Critical roles for transcription factor GATA-3 in thymocyte development. Immunity 19, 863–875 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Wang Y., et al., GATA-3 controls the maintenance and proliferation of T cells downstream of TCR and cytokine signaling. Nat. Immunol. 14, 714–722 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang D.-H., Cohn L., Ray P., Bottomly K., Ray A., Transcription factor GATA-3 is differentially expressed in murine Th1 and Th2 cells and controls Th2-specific expression of the interleukin-5 gene. J. Biol. Chem. 272, 21597–21603 (1997). [DOI] [PubMed] [Google Scholar]
- 11.Zheng W., Flavell R. A., The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 89, 587–596 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Zhu J., et al., Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nat. Immunol. 5, 1157–1165 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Serafini N., et al., Gata3 drives development of RORγt+ group 3 innate lymphoid cells. J. Exp. Med. 211, 199–208 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yagi R., et al., The transcription factor GATA3 is critical for the development of all IL-7Rα-expressing innate lymphoid cells. Immunity 40, 378–388 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhong C., et al., Group 3 innate lymphoid cells continuously require the transcription factor GATA-3 after commitment. Nat. Immunol. 17, 169–178 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoyler T., et al., The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity 37, 634–648 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolterink R. G. J. K., et al., Essential, dose-dependent role for the transcription factor Gata3 in the development of IL-5+ and IL-13+ type 2 innate lymphoid cells. Proc. Natl. Acad. Sci. U.S.A. 110, 10240–10245 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kasal D. N., Bendelac A., Multi-transcription factor reporter mice delineate early precursors to the ILC and LTi lineages. J. Exp. Med. 218, 1–16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong C., et al., Differential expression of the transcription factor GATA3 specifies lineage and functions of innate lymphoid cells. Immunity 52, 83–95.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Constantinides M. G., McDonald B. D., Verhoef P. A., Bendelac A., A committed precursor to innate lymphoid cells. Nature 508, 397–401 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hosoya-Ohmura S., et al., An NK and T cell enhancer lies 280 kilobase pairs 3′ to the gata3 structural gene. Mol. Cell. Biol. 31, 1894–1904 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pandolfi P. P., et al., Targeted disruption of the GATA3 gene causes severe abnormalities in the nervous system and in fetal liver haematopoiesis. Nat. Genet. 11, 40–44 (1995). [DOI] [PubMed] [Google Scholar]
- 23.Lieuw K. H., Li Gl., Zhou Y., Grosveld F., Engel J. D., Temporal and spatial control of murine GATA-3 transcription by promoter-proximal regulatory elements. Dev. Biol. 188, 1–16 (1997). [DOI] [PubMed] [Google Scholar]
- 24.Lakshmanan G., et al., Localization of distant urogenital system-, central nervous system-, and endocardium-specific transcriptional regulatory elements in the GATA-3 locus. Mol. Cell. Biol. 19, 1558–1568 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hasegawa S. L., et al., Dosage-dependent rescue of definitive nephrogenesis by a distant Gata3 enhancer. Dev. Biol. 301, 568–577 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moriguchi T.et al., A Gata3 3′ distal otic vesicle enhancer directs inner ear-specific Gata3 expression. Mol. Cell. Biol. 38, 1–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martynova E., Bouchard M., Musil L. S., Cvekl A., Identification of novel Gata3 distal enhancers active in mouse embryonic lens. Dev. Dyn. 247, 1186–1198 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohmura S., et al., Lineage-affiliated transcription factors bind the Gata3 Tce1 enhancer to mediate lineage-specific programs. J. Clin. Invest. 126, 865–878 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gold M. J., et al., Group 2 innate lymphoid cells facilitate sensitization to local, but not systemic, TH2-inducing allergen exposures. J. Allergy Clin. Immunol. 133, 1142–1148 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Halim T. Y. F., et al., Group 2 innate lymphoid cells are critical for the initiation of adaptive T helper 2 cell-mediated allergic lung inflammation. Immunity 40, 425–435 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yasuda K., Adachi T., Koida A., Nakanishi K., Nematode-infected mice acquire resistance to subsequent infection with unrelated nematode by inducing highly responsive group 2 innate lymphoid cells in the lung. Front. Immunol. 9, 1–15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heng T. S. P., et al., The immunological genome project: Networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Mowel W. K., et al., Group 1 innate lymphoid cell lineage identity is determined by a cis-regulatory element marked by a long non-coding RNA. Immunity 47, 435–449.e8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan W. F., et al., Identification and characterization of the long noncoding RNA Dreg1 as a novel regulator of Gata3. Immunol. Cell Biol. 99, 323–332 (2021). [DOI] [PubMed] [Google Scholar]
- 35.Hu G., et al., Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat. Immunol. 14, 1190–1198 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishizuka I. E., et al., Single-cell analysis defines the divergence between the innate lymphoid cell lineage and lymphoid tissue-inducer cell lineage. Nat. Immunol. 17, 269–276 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hosoya T.et al., Global dynamics of stage-specific transcription factor binding during thymocyte development. Sci. Rep. 8, 1–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo L., et al., IL-1 family members and STAT activators induce cytokine production by Th2, Th17, and Th1 cells. Proc. Natl. Acad. Sci. U.S.A. 106, 13463–13468 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayakawa M., et al., T-helper type 2 cell-specific expression of the ST2 gene is regulated by transcription factor GATA-3. Biochim. Biophys. Acta. 1728, 53–64 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Tindemans I., Serafini N., Di Santo J. P., Hendriks R. W., GATA-3 function in innate and adaptive immunity. Immunity 41, 191–206 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Wei G., et al., Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity 35, 299–311 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu J., Yamane H., Paul W. E., Differentiation of effector CD4 T cell populations (*). Annu. Rev. Immunol. 28, 445–489 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Furusawa J., et al., Critical role of p38 and GATA3 in natural helper cell function. J. Immunol. 191, 1818–1826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Molofsky A. B., et al., Innate lymphoid type 2 cells sustain visceral adipose tissue eosinophils and alternatively activated macrophages. J. Exp. Med. 210, 535–549 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nussbaum J. C., et al., Type 2 innate lymphoid cells control eosinophil homeostasis. Nature 502, 245–248 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verhoef P. A., et al., Intrinsic functional defects of type 2 innate lymphoid cells impair innate allergic inflammation in promyelocytic leukemia zinc finger (PLZF)-deficient mice. J. Allergy Clin. Immunol. 137, 591–600.e1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li B. W. S., Beerens D. M. J. M., Brem M. D., Hendriks R. W., “Characterization of Group 2 Innate Lymphoid Cells in Allergic Airway Inflammation Models in the Mouse” in Inflammation, Methods in Molecular Biology, Clausen B., Laman J., Eds. (Humana Press, New York, NY, 2017), 1559. [DOI] [PubMed] [Google Scholar]
- 48.Cipolletta D., et al., PPAR-γ is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature 486, 549–553 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han J. M., et al., IL-33 reverses an obesity-induced deficit in visceral adipose tissue ST2+ T regulatory cells and ameliorates adipose tissue inflammation and insulin resistance. J. Immunol. 194, 4777–4783 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Dahlgren M. W., et al., Adventitial stromal cells define group 2 innate lymphoid cell tissue niches. Immunity 50, 707–722.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spencer S. P., et al., Adaptation of innate lymphoid cells to a micronutrient deficiency promotes type 2 barrier immunity. Science 343, 432–437 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mukai K., Karasuyama H., Kabashima K., Kubo M., Galli S. J., Differences in the importance of mast cells, basophils, IgE, and IgG versus that of CD4+ T cells and ILC2 cells in primary and secondary immunity to Strongyloides venezuelensis. Infect. Immun. 85, 1–18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yasuda K., et al., Contribution of IL-33-activated type II innate lymphoid cells to pulmonary eosinophilia in intestinal nematode-infected mice. Proc. Natl. Acad. Sci. U.S.A. 109, 3451–3456 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fang D., et al., Bcl11b, a novel GATA3-interacting protein, suppresses Th1 while limiting Th2 cell differentiation. J. Exp. Med. 215, 1449–1462 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miyamoto C., et al., Runx/Cbfβ complexes protect group 2 innate lymphoid cells from exhausted-like hyporesponsiveness during allergic airway inflammation. Nat. Commun. 10, 1–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hosokawa H.et al., Cell type-specific actions of Bcl11b in early T-lineage and group 2 innate lymphoid cells. J. Exp. Med. 217, 1–14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spooner C. J., et al., Specification of type 2 innate lymphocytes by the transcriptional determinant Gfi1. Nat. Immunol. 14, 1229–1236 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Wei L., et al., Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity 32, 840–851 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wingender E., The TRANSFAC project as an example of framework technology that supports the analysis of genomic regulation. Brief. Bioinform. 9, 326–332 (2008). [DOI] [PubMed] [Google Scholar]
- 60.Ouyang W., et al., Stat6-independent GATA-3 autoactivation directs IL-4-independent Th2 development and commitment. Immunity 12, 27–37 (2000). [DOI] [PubMed] [Google Scholar]
- 61.Zhu J., T helper 2 (Th2) cell differentiation, type 2 innate lymphoid cell (ILC2) development and regulation of interleukin-4 (IL-4) and IL-13 production. Cytokine 75, 14–24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barolo S., Shadow enhancers: Frequently asked questions about distributed cis-regulatory information and enhancer redundancy. BioEssays 34, 135–141 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guenther C. A., Tasic B., Luo L., Bedell M. A., Kingsley D. M., A molecular basis for classic blond hair color in Europeans. Nat. Genet. 46, 748–752 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Naik S., et al., Inflammatory memory sensitizes skin epithelial stem cells to tissue damage. Nature 550, 475–480 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Poulin F., et al., In vivo characterization of a vertebrate ultraconserved enhancer. Genomics 85, 774–781 (2005). [DOI] [PubMed] [Google Scholar]
- 66.Snider P., et al., Generation and characterization of Csrp1 enhancer‐driven tissue‐restricted Cre‐recombinase mice. genesis 46, 167–176 (2008). [DOI] [PubMed] [Google Scholar]
- 67.Tasic B., et al., Site-specific integrase-mediated transgenesis in mice via pronuclear injection. Proc. Natl. Acad. Sci. U.S.A. 108, 7902–7907 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kurata H., Lee H. J., O’Garra A., Arai N., Ectopic expression of activated Stat6 induces the expression of Th2-specific cytokines and transcription factors in developing Th1 cells. Immunity 11, 677–688 (1999). [DOI] [PubMed] [Google Scholar]
- 69.Zhu J., Guo L., Watson C. J., Hu-Li J., Paul W. E., Stat6 is necessary and sufficient for IL-4's role in Th2 differentiation and cell expansion. J. Immunol. 166, 7276–7281 (2001). [DOI] [PubMed] [Google Scholar]
- 70.Ishizuka I. E., Constantinides M. G., Gudjonson H., Bendelac A., The innate lymphoid cell precursor. Annu. Rev. Immunol. 34, 299–316 (2016). [DOI] [PubMed] [Google Scholar]
- 71.Stabile H., et al., JAK/STAT signaling in regulation of innate lymphoid cells: The gods before the guardians. Immunol. Rev. 286, 148–159 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Halim T. Y. F., et al., Group 2 innate lymphoid cells license dendritic cells to potentiate memory TH 2 cell responses. Nat. Immunol. 17, 57–64 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Noben-Trauth N., Hu-Li J., Paul W. E., IL-4 secreted from individual naive CD4+ T cells acts in an autocrine manner to induce Th2 differentiation. Eur. J. Immunol. 32, 1428–1433 (2002). [DOI] [PubMed] [Google Scholar]
- 74.Corces M. R., et al., An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Buenrostro J. D., Wu B., Chang H. Y., Greenleaf W. J., ATAC‐seq: A method for assaying chromatin accessibility genome‐wide. Curr. Protoc. Mol. Biol. 109, 21–29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Heinz S., et al., Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kasal D. N., Bendelac A., A Gata3 enhancer necessary for ILC2 development and function. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE169542. Deposited 24 March 2021. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ATAC-seq data have been deposited in Gene Expression Omnibus (GSE169542) (77). All other study data are included in the article and/or supporting information.