Figure 3.
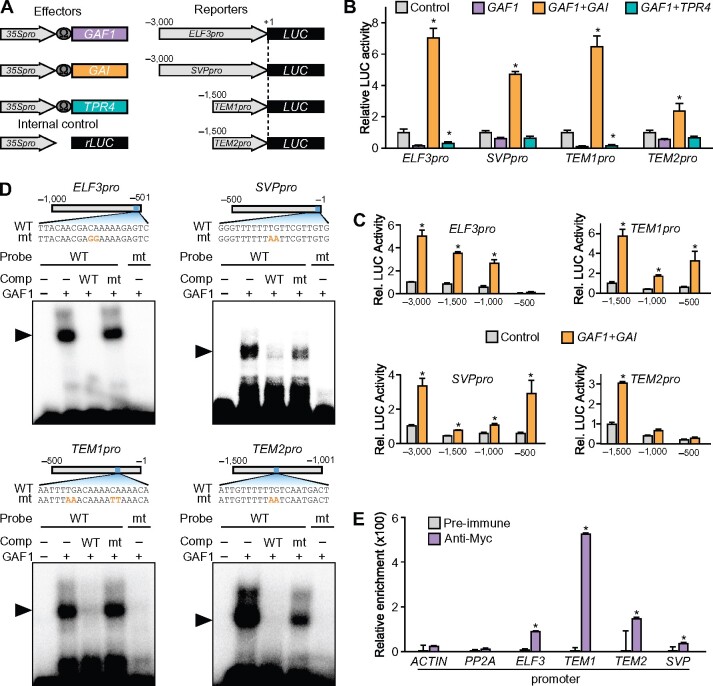
Identification of novel GAF1 target genes involved in the regulation of flowering. A, Schematic representation of the reporter and effector. Fragments (3,000 bp) of the ELF3 and SVP promoters and 1,500-bp fragments of the TEM1 and TEM2 promoters were fused with the LUC gene. The effector plasmid expressed full-length GAF1, GAI, or TPR4 under control of the CaMV 35S promoter with a viral translation enhancer (Ω). B, Transient expression assay showed that the DELLA-GAF1 complex activated and the TPR4-GAF1 complex repressed the ELF3, SVP, TME1, and TEM2 promoters. The reporter plasmids consisted of the 3,000-bp promoter regions of ELF3 and SVP and the 1,500-bp promoter regions of TEM1 and TEM2 fused with the LUC reporter gene. The results are shown as LUC/rLUC activity. Error bars indicate the sd of three biological replicates (n = 3). Asterisks represent Student’s t test significance compared with mock treated control (*P < 0.05). C, Transactivation assay of GAF1 and GAI. The effector, reporter, and internal control constructs were co-transfected into Arabidopsis protoplasts. The transfected cells were incubated for 20 h, and Luc and rLUC activities were measured. The results are shown as LUC/rLUC activity. Error bars indicate sd of the mean (n = 3). Asterisks represent Student’s t test significance compared with mock treated control (*P < 0.05). D, Identification of GAF1-binding regions in the ELF3, SVP, TEM1, and TEM2 promoters in vitro. EMSA analysis using recombinant GAF1 protein. Oligonucleotides containing ELF3pro (−529 to −510, wild-type; lanes 1–4) or mutated (mt) ELF3pro (mt; lane 5), SVPpro (−20 to −1, wild-type; lanes 1–4) or mtSVPpro (mt; lane 5), TEM1pro (−152 to −133, wild-type; lanes 1–4) or mtTEM1pro (mt; lane 5), and TEM2pro (−1,288 to −1,269, wild-type; lanes 1–4) or mtTEM2pro (mt; lane 5) were used as probes. Orange letters indicate mutated bases. Wild-type and mt indicate competition with a 100-fold to 500-fold excess of unlabeled wild-type and mutated probe, respectively. The specific GAF1–DNA complexes are indicated by arrowheads. +, Addition to the reaction mixtures; –, omission from the reaction mixtures. E, Plants were grown under SD conditions for 2 weeks. Whole seedlings were harvested at ZT8. GAF1 binds to a region of the ELF3, SVP, TEM1, and TEM2 promoters in vivo. ChIP assays were performed with pre-immune or anti-myc in CaMV 35S:myc-GAF1 transgenic plants. The co-precipitated level of each DNA fragment was quantified by real-time PCR and normalized to the input DNA. Error bars indicate sd of three technical replicates (n = 3). Asterisks represent Student’s t test significance compared with pre-immune as control (*P < 0.05). Experiments were repeated twice with independently grown plants, with similar results.