Abstract
Extreme temperature conditions seriously impair male reproductive development in plants; however, the molecular mechanisms underlying the response of anthers to extreme temperatures remain poorly described. The transcription factor phytochrome-interacting factor4 (PIF4) acts as a hub that integrates multiple signaling pathways to regulate thermosensory growth and architectural adaptation in plants. Here, we report that SlPIF4 in tomato (Solanum lycopersicum) plays a pivotal role in regulating cold tolerance in anthers. CRISPR (clustered regularly interspaced short palindromic repeats)–associated nuclease Cas9-generated SlPIF4 knockout mutants showed enhanced cold tolerance in pollen due to reduced temperature sensitivity of the tapetum, while overexpressing SlPIF4 conferred pollen abortion by delaying tapetal programmed cell death (PCD). SlPIF4 directly interacts with SlDYT1, a direct upstream regulator of SlTDF1, both of which (SlDYT1 and SlTDF1) play important roles in regulating tapetum development and tapetal PCD. Moderately low temperature (MLT) promotes the transcriptional activation of SlTDF1 by the SlPIF4–SlDYT1 complex, resulting in pollen abortion, while knocking out SlPIF4 blocked the MLT-induced activation of SlTDF1. Furthermore, SlPIF4 directly binds to the canonical E-box sequence in the SlDYT1 promoter. Collectively, these findings suggest that SlPIF4 negatively regulates cold tolerance in anthers by directly interacting with the tapetal regulatory module in a temperature-dependent manner. Our results shed light on the molecular mechanisms underlying the adaptation of anthers to low temperatures.
The transcription factor PIF4 negatively regulates cold tolerance in tomato anthers by directly interacting with the tapetal regulatory module DYT1-TDF1-AMS in a temperature-dependent manner.
INTRODUCTION
Fruits and seeds, which constitute a major component of the human diet, are mainly derived from the sexual reproduction of flowering plants. However, plants are at the mercy of environmental factors, including temperature, which can significantly affect the fertilization process and result in failed fruit set (Wang et al., 2009; Hatfield and Prueger, 2015). In this regard, there is growing concern about the effects of global climate change, and extreme temperature events have already had a widespread impact on plant pollination and fertilization, resulting in a substantial reduction in crop yield (Lobell and Gourdji, 2012). Extreme temperatures can affect both male and female development in flowering plants, and pollen and anther development are particularly sensitive to ambient temperature fluctuations (Hedhly et al., 2009; Thakur et al., 2010; Müller and Rieu, 2016; Smith, 2019). However, the molecular mechanisms underlying temperature stress tolerance are still poorly understood. An improved understanding of the basis of temperature-related male sterility would be important for sustaining crop production and enhancing food security.
Phytochrome-interacting factors (PIFs), which comprise a small subset of basic–helix–loop–helix (bHLH) family transcription factors, act as central regulators in the integration of light and temperature signals to optimize plant growth and development (Castillon et al., 2007; Leivar and Quail, 2011). To date, eight PIF genes (PIF1–PIF8), belonging to bHLH subgroup 15, have been identified and characterized in Arabidopsis thaliana (Toledo-ortiz et al., 2003; Lee and Choi, 2017). PIF4 acts as a central hub that integrates signals through circadian, light, ambient temperature, phytohormonal, and defense-associated pathways to modulate a range of adaptive growth processes in plants (Paik et al., 2017). These processes include the regulation of plant adaptation to temperature stress, thermo-morphogenic phenotypes (e.g. hyponasty, hypocotyl, and petiole lengths), and flowering time (Quint et al., 2016). PIF4 functions in a tissue-specific manner during thermo-morphogenesis, although the gene is expressed in most tissues (Kim et al., 2020). Furthermore, by interacting with PIF7 and phytochrome B (PHYB), PIF4 negatively regulates the low-temperature tolerance of A. thaliana seedlings by diurnally repressing the C-repeat binding factor (CBF) pathway (Catalá et al., 2011; Lee and Thomashow, 2012). PIF3 functions as a negative regulator of freezing tolerance in Arabidopsis by integrating light and low-temperature signaling (Jiang et al., 2017; Lin et al., 2018). However, the role of PIFs in regulating the adaptation of reproductive organs to temperature stress is unclear.
Research into male reproductive development over the last decade has revealed that sugar, phytohormone, and reactive oxygen species (ROS) signaling pathways play important roles in plant responses to temperature stress (Giorno et al., 2013; Min et al., 2014; Rieu et al., 2017). In anthers, temperature stress disturbs the normal functions of the tapetum (Zinn et al., 2010; De Storme and Geelen, 2014), the innermost cell layer of the anther. The tapetum surrounds the microspores and supplies enzymes, nutrients, and precursors for microspore and pollen wall development as a consequence of programmed cell death (PCD)-triggered tapetal degradation (Ma, 2005; Ariizumi and Toriyama, 2011; Xiong et al., 2016). This tapetum-specific PCD is essential for proper microspore development and pollen maturation. Numerous reports have ascribed the induction of male sterility to premature or delayed degradation of the tapetum (Vizcay-Barrena and Wilson, 2006; Liu and Fan, 2013; Shi et al., 2015). The tapetum is especially sensitive to environmental stress at the young microspore stage (Müller and Rieu, 2016), and in A. thaliana (Baron et al., 2012), rice (Oryza sativa; Ku et al., 2003), wheat (Triticum aestivum; Omidi et al., 2014), and barley (Hordeum vulgare; Oshino et al., 2007), high temperature induces premature degeneration of the tapetal cell layer. In contrast, cold stress delays or inhibits tapetum regeneration by disrupting tapetal PCD (Sharma and Nayyar, 2016; Liu et al., 2019; Xu et al., 2020). Studies using forward genetic approaches have indicated that a genetic module composed of Dysfunctional Tapetum 1 (DYT1), Defective in Tapetal Development and Function 1 (TDF1), Aborted Microspores (AMS), and MYB80 regulates tapetum development and tapetal PCD and that this module is evolutionarily conserved in A. thaliana, rice, maize (Zea mays), tomato (Solanum lycopersicum), and other crops (Jung et al., 2005; Li et al., 2006; Phan et al., 2011; Niu et al., 2013; Ko et al., 2014; Jeong et al., 2014; Li et al., 2017, Nan et al., 2017). The leucine-rich repeat-receptor-like kinases TMS10 and TMS10L regulate tapetal degeneration in rice under fluctuating temperatures (Yu et al., 2017), and the DICER-LIKE (DCL) protein Del5 is essential for maintaining male fertility and tapetal development under high temperature in maize (Teng et al. 2020). However, the molecular link between environmental stress and male reproductive development in flowering plants is currently unclear.
A potentially valuable system for investigating the roles of PIF proteins in regulating plant fertility under temperature stress is tomato, one of the most widely cultivated vegetable crops worldwide. Eight PIF family members (SlPIF1a, SlPIF1b, SlPIF3, SlPIF4, SlPIF7a, SlPIF7b, SlPIF8a, and SlPIF8b) have been identified in the tomato genome (Rosado et al., 2016). SlPIF4 positively regulates cold stress tolerance in tomato seedlings by directly activating the expression of CBF genes (Wang et al., 2020), suggesting that PIF4 might have undergone functional diversification between different plant species.
In this study, we characterized the role of SlPIF4 in regulating the adaptation of anthers to moderately low temperature (MLT) stress by characterizing CRISPR/Cas9-mediated SlPIF4-knockout mutants (slpif4) and SlPIF4-overexpressing (OE) lines. We discovered that slpif4 anthers exhibited enhanced cold tolerance due to altered tapetal PCD, while overexpression of SlPIF4 caused pollen abortion due to delayed tapetal PCD. We further demonstrated that SlPIF4 physically interacts with SlDYT1 and directly activates SlDYT1 expression. Moreover, MLT promotes the transcriptional activation of SlTDF1 by the SlPIF4–SlDYT1 complex, thereby postponing tapetal PCD, while this activation is completely inhibited in slpif4 anthers under MLT stress. These results indicate that SlPIF4 mediates anther development and cold tolerance in a temperature-dependent manner. Our study sheds light on the molecular mechanisms underlying the adaptation of anthers to temperature stress.
RESULTS
SlPIF4 is expressed in tomato flowers in a photoperiod-independent manner
Similar to A. thaliana PIF proteins, tomato SlPIF4 contains a conserved active phytochrome B-binding (APB) domain and a bHLH DNA-binding domain, although a conserved Q-residue is substituted with an E-residue in the APB-binding domain of SlPIF4 (Supplemental Figure S1). However, SlPIF4 only shares 40% and 41% amino acid sequence identity with its two corresponding homologs in A. thaliana, AtPIF4 and AtPIF5, respectively (Supplemental Figure S1). Subcellular localization analysis via heterologous expression of a SlPIF4-green fluorescent protein (GFP) fusion protein in Nicotiana benthamiana epidermal cells revealed a fluorescent signal exclusively located in the cell nucleus (Supplemental Figure S2), indicating SlPIF4 is a nucleus-localized protein.
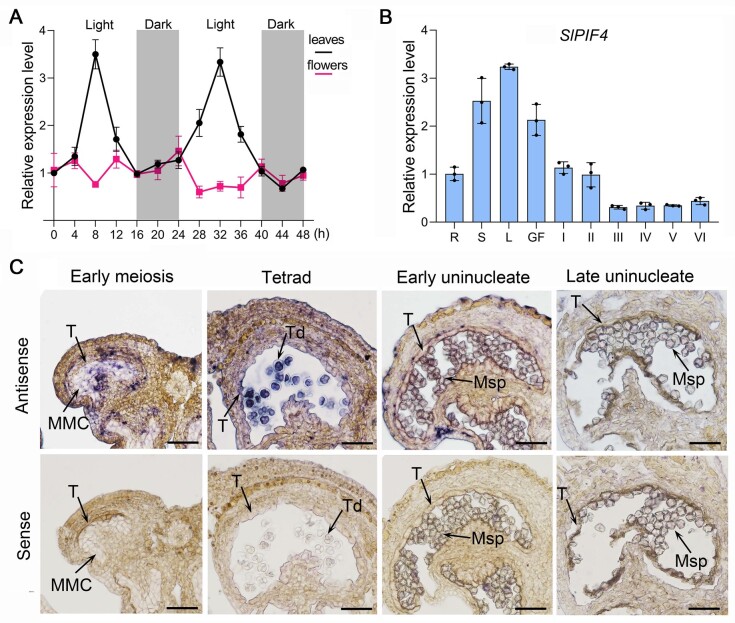
The expression profiles of PIF genes generally show diurnal oscillations in vegetative organs, including leaves and stems (Jeong et al., 2014). Here, we found that SlPIF4 was expressed in a circadian manner in the leaves of tomato plants grown under a 16-h light/8-h dark photoperiod, with peaks in the middle of the light period, while during the dark period, the transcripts gradually accumulated from midnight to dawn (Figure 1A). The SlPIF4 expression pattern in tomato leaves is consistent with that of PIF4 in A. thaliana and rice (Todaka et al., 2012; Yamashino et al., 2013). In tomato flower buds, however, the expression levels of SlPIF4 barely changed during the same photoperiodic regime (Figure 1A), suggesting that SlPIF4 is not circadian-regulated in flowers.
Figure 1.
Expression analysis of SlPIF4. A, Expression profile of SlPIF4 in tomato leaves and flowers. “Micro-Tom” seedlings were grown under long-day conditions (16-h/8-h light/dark photoperiod) for 6 weeks, and RNA samples were prepared from the third fully expanded leaf and flower buds at the binucleate stage at a 4-h interval. Three individual plants were used for sampling at each time point. 0 h: 6 a.m. The dark periods are indicated by gray shading. Each value is the mean ± sd (n = three biological replicates). Expression levels shown are relative to that at time point 0, which was set at 1. B and C, Expression analysis of SlPIF4 in various tissues using qRT-PCR (B) and in WT anthers by in situ hybridization (C). Each value indicates the mean ± sd (n = three technical replicates, each organ was selected from at least five individual WT plants). Expression levels shown in (B) are relative to that in roots, which was set at 1. R, roots; S, stems; L, leaves; GF, green fruits; I, early meiosis stage; II, tetrad stage; III, early uninucleate stage; IV, late uninucleate stage; V, binucleate stage; VI, mature pollen stage. MMC, microspore mother cell; Msp, microspore; T, tapetum; Td, tetrads. Bars = 100 µm.
SlPIF4 transcripts primarily accumulated in leaves, stems, and green fruits (Figure 1B) but were also detected in other organs. During anther development, SlPIF4 expression was high at Stages I (early meiosis) and II (tetrad), and showed a significant decrease at Stage III (early uninucleate), and remained at a low level during later stages (Figure 1B). In situ hybridization analysis showed that SlPIF4 was mainly present in microspore mother cells at Stage I, and in tapetal cells and microspores at Stage II, but there was a minimal signal from Stage III onward (Figure 1C). These results suggest that SlPIF4 might play a role during early anther development.
Knocking out SlPIF4 enhances the tolerance of anthers to moderate high- and low-temperature stress
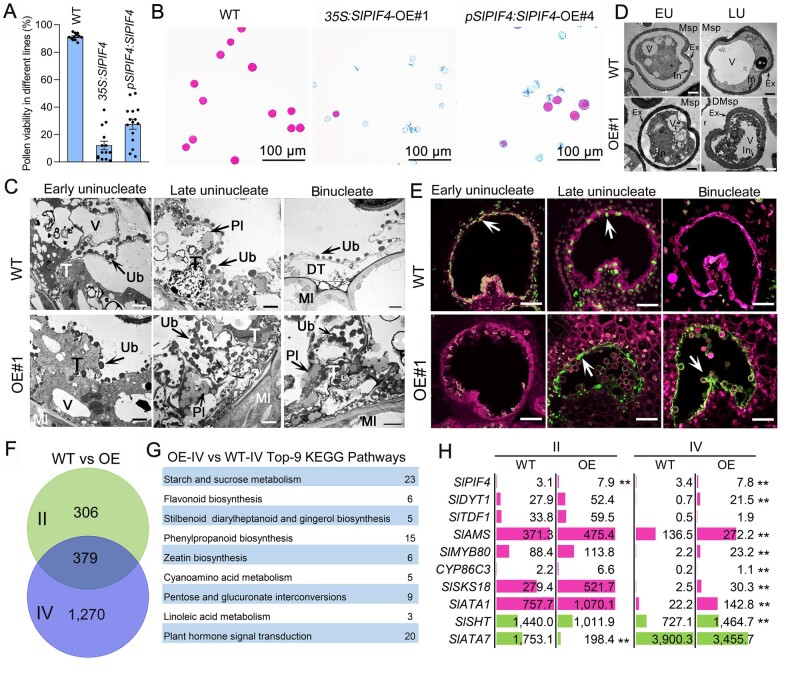
To characterize the function of SlPIF4, we generated SlPIF4-knockout mutants via CRISPR/Cas9 gene editing with guide RNA targeting the bHLH DNA-binding domain (Supplemental Figure S3, A). Two slpif4 homozygous mutant lines (slpif4-13 and slpif4-9) were subjected to further analysis. slpif4-13 was found to contain a 1-bp deletion that caused a frameshift mutation in the SlPIF4 coding sequence, leading to a pre-termination of the bHLH domain, while the edit in slpif4-9 was predicted to result in one amino acid substitution and a two amino acid deletion in the bHLH domain (Supplemental Figure S3A). Both slpif4 mutants produced viable pollen with normal fruit set (Supplemental Figure S3, B and C), indicating that the loss of SlPIF4 function does not affect fertility under normal growth conditions.
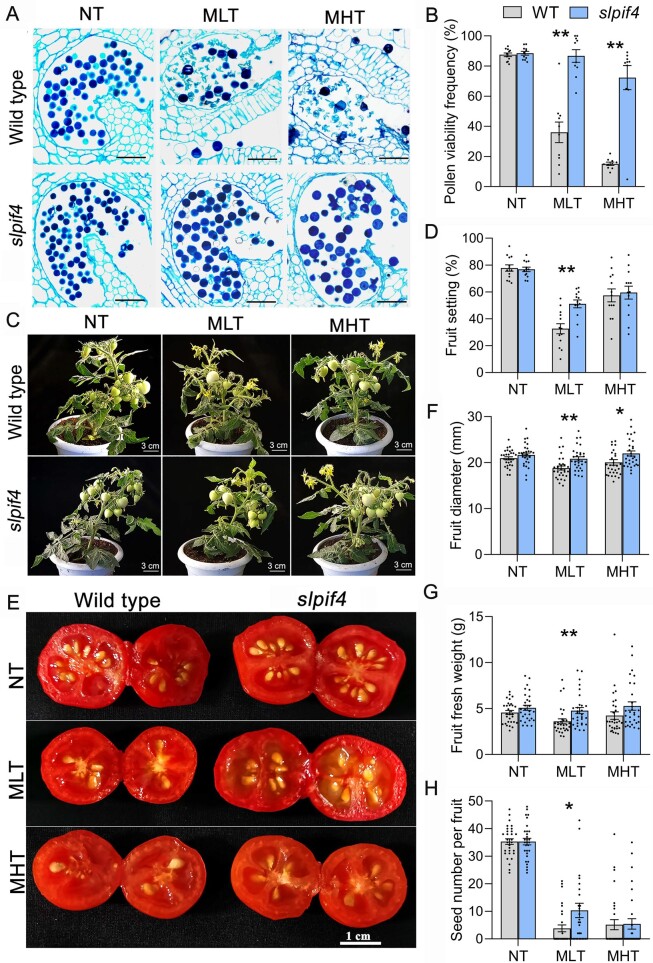
Given the importance of PIF4 in regulating plant adaptation and the susceptibility of male gametophyte development to ambient temperature stress, we compared anther development in wild-type (WT) plants and slpif4 mutants grown under a moderately high temperature (MHT) regime (32°C/27°C, day/night) for 4 d or a MLT regime (15°C/15°C, day/night) for 6 d. During growth under normal temperature (NT) conditions (25°C/20°C, day/night), WT and slpif4 plants showed high pollen viability (Figure 2, A and B), while only 35.9% and 15.1% of the pollen grains were viable in WT plants after MLT and MHT treatment, respectively (Figure 2, A and B). However, the number of viable pollen grains increased to 86.7% and 72.3% in slpif4 mutants grown under MLT and MHT, respectively (Figure 2, A and B). As expected, MLT and MHT treatments significantly reduced the fruit set from 78.8% to 32.6% and 57.4%, respectively, in WT plants (Figure 2, C and D), whereas the fruit set rate of the slpif4 mutants only decreased to 51.1% compared to 32.6% in WT plants under MLT (Figure 2, C and D).
Figure 2.
Knocking out SlPIF4 enhances tolerance of tomato pollen to moderate temperature stress. A, C, and E, Representative images of semithin sections of anthers (A), plants during fruiting periods (C), and transverse sections of red fruits in WT and slpif4 plants under NT, MLT, and MHT conditions. Five to 10 anthers at the tetrad stage from the 6-week-old plants were randomly marked with strings for each plant before temperature treatments. The marked anthers were examined for pollen viability (A) at the mature pollen stage and fruit traits at the breaker (C) and ripening (E) stages of fruits after exposure to temperature stress. MHT, 32°C/28°C, day/night for 4 d; MLT, 15°C/15°C, day/night for 6 d; NT, 25°C/20°C, day/night. B, D, F, G, and H, Determination of pollen viability (B), fruit setting rate (D), fruit diameter (F), fruit fresh weight (G), and seed number per fruit (H) in WT and slpif4 plants grown under NT, MLT, and MHT conditions. Each black dot represents the value of pollen viability per pollen sac fruit setting rate per plant (D), fruit diameter (F), fruit fresh weight (G), and seed number (H) per red fruit. Each error bar represents the mean ± se, n = 8–12 pollen sacs in (B), 12 plants in (D), 30 red fruits in (F), (G), and (H). Asterisks indicate significant differences: *P < 0.05, **P < 0.01 (Student’s t test). The data for fruit diameter, fresh fruit weight, and number of seeds per fruit were subjected to normality tests using both D'Agostino–Pearson and Anderson–Darling tests, and all data passed the test (alpha = 0.05). Bars in (A) = 50 µm.
We then compared fruit and seed development in WT and slpif4 plants grown under NT, MHT, and MLT conditions. Compared to WT plants, no significant difference was detected in fruit diameter, fresh fruit weight, or number of seeds per fruit in slpif4 mutants under NT (Figure 2, E–H). Rosado et al. (2019) reported that SlPIF4-silenced individual red fruits were smaller in diameter compared to the WT. We speculate that the difference in fruit size between these studies might be due to differences in the methods used for genetic silencing or the different cultivation conditions. In contrast, significantly larger fruit diameters were observed in slpif4 under MLT or MHT treatment (Figure 2F). Correspondingly, the fruit weight of slpif4 was consistently higher than that of WT plants after either of the two treatments, particularly MLT (Figure 2G). Furthermore, under MLT conditions, the seed number per fruit in slpif4 was 1.7-fold greater than that in WT plants (Figure 2H). In addition, slpif4 anthers produced significantly more viable pollen grains than WT anthers following treatment at 10°C for 6 d (Supplemental Figure S4A). These results indicate that SlPIF4 plays an important role in yield production in tomatoes under cold stress. Thus, we focused on exploring the mechanism underlying the SlPIF4-regulated low-temperature tolerance of tomato anthers.
SlPIF4 negatively regulates MLT tolerance of tomato anthers by regulating tapetal PCD
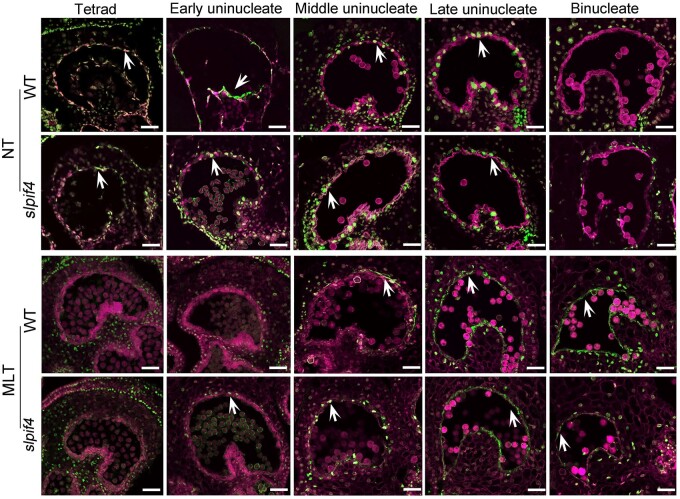
The timing of PCD-mediated degeneration of tapetal cells is critical for pollen development (Falasca et al., 2013). To determine the timing of tapetal PCD in WT and slpif4 anthers, we performed a terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay using tomato anthers at different developmental stages from plants grown under NT or MLT conditions. WT and slpif4 anthers showed a similar pattern of tapetal PCD under NT conditions (Figure 3). TUNEL-positive signals first appeared in tapetal cells at the tetrad stage, intensified from the early to late uninucleate stage, and were absent at the binucleate stage in both WT and slpif4 anthers under NT conditions (Figure 3). However, after MLT treatment, the tapetal PCD of WT anthers was significantly delayed, as TUNEL-positive signals were not detected until the middle uninucleate stage and were intense at the late uninucleate and even the binucleate stage (Figure 3). In contrast, in slpif4 anthers, the delayed tapetal PCD caused by MLT treatment was alleviated, as a weak TUNEL-positive signal appeared at the early uninucleate stage, became intense at the middle, and late uninucleate stage, and weakened at the binucleate stage (Figure 3). In addition, we compared the morphology of WT and slpif4 anther tapetum at the tetrad stage by transmission electron microscopy (Supplemental Figure S4B). MLT treatment considerably reduced the number of vacuoles and mitochondria in the tapetum, particularly for WT anthers (Supplemental Figure S4B). In addition, the tapetum remained much more intact and densely stained after MLT treatment, which is consistent with the finding (from TUNEL assays) that tapetal PCD was delayed in tomato anthers by MLT treatment (Figure 3).
Figure 3.
Knocking out SlPIF4 reduces the sensitivity of the tapetum to moderately low-temperature conditions. TUNEL assays of tapetal PCD in WT and slpif4 anthers under NT and MLT conditions at the tetrad, early, middle and late uninucleate, and late binucleate stages. The magenta signal is propidium iodide staining, and the green fluorescence indicates TUNEL positive signal (white arrows). At least seven anthers in each stage were sampled for TUNEL assays after 6-week-old WT and slpif4 plants were exposed to MLT (15°C/15°C, day/night) and NT (25°C/20°C, day/night) for 6 d. Bars = 50 µm.
Knocking out SlPIF4 blocks MLT-induced activation of SlTDF1
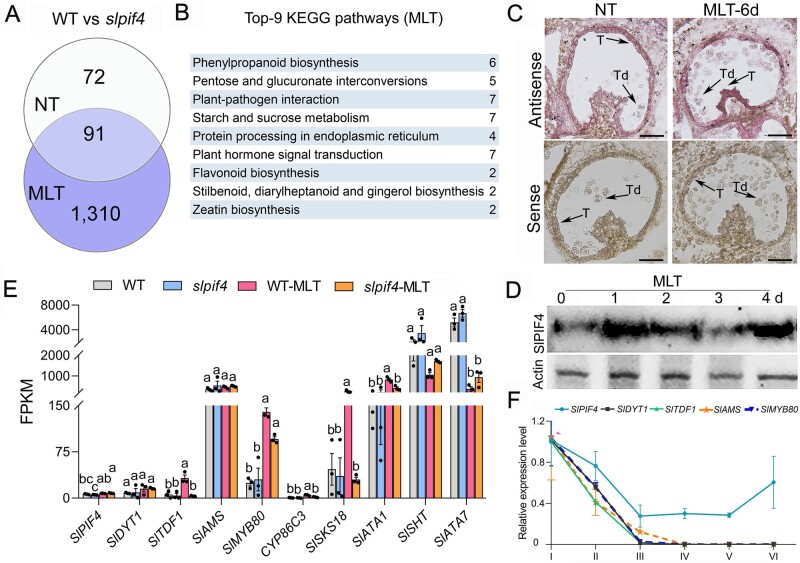
To further characterize the role of SlPIF4 in regulating MLT tolerance in anthers, we compared the transcriptome profiles of WT and slpif4 anthers at the early uninucleate stage following NT and MLT treatment. A comparison of expression profiles revealed high Pearson correlation coefficients between the three biological replicates (Supplemental Figure S5, A and B), pointing to the high reproducibility of the transcriptome data. A total of 4,385 and 3,613 differentially expressed genes (DEGs) were identified in WT and slpif4 anthers under MLT conditions, respectively (Supplemental Figure S5C and Supplemental Data Sets S1 and S2). In WT anthers, 2,684 and 1,701 genes were up- and downregulated, respectively, and in slpif4 anthers, 1,969 and 1,644 genes were up- and downregulated, respectively. Notably, a total of 1,416 DEGs were uniquely identified in slpif4 anthers under MLT treatment (Supplemental Figure S5C). Interestingly, under NT conditions, only 163 DEGs were identified between slpif4 and WT anthers (Figure 4A;Supplemental Data Set S3). However, the number of DEGs between slpif4 and WT anthers markedly increased to 1,401 after MLT treatment, with 920 genes upregulated and 481 downregulated in slpif4 anthers (Figure 4A).
Figure 4.
MLT conditions cause expression changes of genes regulating tapetum and pollen development in WT and slpif4 anthers. A and B, Venn diagram (A) and KEGG analysis (B) of DEGs between WT and slpif4 anthers at the early uninucleate stage grown under MLT or NT conditions. C, In situ hybridization analysis of SlPIF4 mRNA in WT anthers at the tetrad stage under NT and MLT conditions. At least six anthers at the tetrad stage were sampled for in situ hybridization assays after 6-week-old WT and slpif4 plants were exposed to MLT (15°C/15°C, day/night) and NT (25°C/20°C, day/night) for 6 d. T, tapetum; Td, tetrad. Bars = 100 µm. D, SlPIF4 protein levels in WT anthers at stages II and III at 0, 24, 48, 72, and 96 h post-MLT treatment. II, tetrad stage; III, early uninucleate stage; d, day. E, DEGs involved in tapetum and pollen wall development between WT and slpif4 anthers under NT and MLT conditions. Values represent gene expression levels (FPKM) based on RNA-Seq data. Each error bar represents the mean ± se, n = three biological replicates. Different letters indicate significantly different mean values at P < 0.05 (one-way ANOVA with posthoc Tukey test). F, Expression analysis of SlPIF4, SlDYT1, SlTDF1, SlAMS, and SlMYB80 in WT anthers at Stages I–VI by qRT-PCR. I, early meiosis stage; II, tetrad stage; III, early uninucleate stage; IV, late uninucleate stage; V, binucleate stage; VI, mature pollen stage. At least 10 anthers in each stage were sampled from 10 individual plants for each biological replicate. Each error bar represents the mean ± sd, n = three biological replicates.
KEGG (Kyoto Encyclopedia of Genes and Genomes) analysis revealed that the aforementioned 1,401 DEGs were significantly enriched in the pathways phenylpropanoid biosynthesis, metabolism of starch and sucrose, and plant hormone signal transduction (Figure 4B). Thus, we measured the endogeno us indoleacetic acid (IAA), jasmonic acid (JA), and abscisic acid (ABA) levels in both WT and slpif4 anthers at the tetrad stage by HPLC-mass spectrometry. Under NT conditions, compared to WT anthers, in slpif4 anthers, the endogenous IAA contents decreased by 24% and the endogenous ABA contents increased by 86% (Supplemental Figure S6). However, under MLT conditions, only the endogenous JA contents significantly increased in slpif4 anthers, while neither IAA nor ABA contents exhibited significant differences between WT and slpif4 anthers (Supplemental Figure S6).
Based on the RNA-Seq data, the expression of SlPIF4 in WT anthers increased by 18.6% under MLT conditions (Figure 4E). In situ hybridization revealed that SlPIF4 mRNA strongly accumulated in tapetal cells under MLT conditions (Figure 4C). Furthermore, immunoblot analysis showed that the abundance of SlPIF4 in WT anthers increased following MLT treatment (Figure 4D). In addition, the transcriptional DYT1-TDF1-AMS-MYB80 cascade, which modulates tapetal PCD, was induced by MLT in WT anthers (Figure 4E). The expression level of SlDYT1 increased by 83% under MLT conditions (Figure 4E), and SlTDF1 and SlMYB80 were significantly upregulated (Figure 4E). In addition, multiple downstream genes regulated by this transcriptional cascade, including CYP86C3, SlSKS18, and SlATA1, were significantly upregulated in WT anthers by MLT treatment (Figure 4E;Supplemental Figure S5D). Two genes that participate in pollen wall development, SlSHT and SlATA7, were substantially downregulated by MLT in WT anthers (Figure 4E).
In contrast, in slpif4 anthers, among the transcriptional DYT1-TDF1-AMS-MYB80 module, the expression of SlDYT1 was induced by MLT to the same high level as in WT anthers following MLT treatment (Figure 4E). Notably, the expression of SlTDF1 was not induced by MLT treatment in slpif4 anthers (Figure 4E). SlMYB80, the downstream gene of SlTDF1, was less responsive to MLT in slpif4 anthers than the WT, and another downstream gene, SlAMS, displayed almost no change in expression after MLT treatment (Figure 4E). Furthermore, multiple downstream genes of the DYT1-TDF1-AMS cascade, including CYP86C3, SlSKS18, and SlATA1 showed strikingly less change in expression under MLT treatment in slpif4 anthers compared to WT anthers (Figure 4E;Supplemental Figure S5D). Two-way analysis of variance (ANOVA) further revealed that SlPIF4 alone had a significant effect on the expression of genes related to tapetal PCD, particularly SlTDF1 (Supplemental Figure S5E), indicating that the differential response of WT and slpif4 anthers to MLT could be attributed to SlPIF4. Taken together, these results suggest that SlPIF4 is required for the activation of SlTDF1 under MLT conditions.
SlPIF4 had a similar expression pattern to SlDYT1, SlTDF1, SlAMS, and SlMYB80 in WT anthers at Stages I–III, with a rapid decrease in transcript abundance through these three stages (Figure 4F). This result further supports the notion that SlPIF4 acts together with the transcriptional DYT1-TDF1-AMS-MYB80 cascade in response to low temperature during the early stages of tapetum development.
SlPIF4 overexpression results in delayed tapetal PCD and pollen infertility
To further investigate the role of SlPIF4 in anther development, we overexpressed the SlPIF4 cDNA sequence under the control of the constitutive cauliflower mosaic virus 35S promoter or its native promoter pSlPIF4. Thirteen independent 35S:SlPIF4 and 14 pSlPIF4:SlPIF4 transgenic lines were obtained, all of which displayed normal flowering (Supplemental Figure S7). However, under NT conditions, the pollen viability of the 35S:SlPIF4 and pSlPIF4:SlPIF4 lines was reduced to 1.4%–38.3% and 3.8%–49.9%, respectively, compared to the average value of 91.1% in WT plants (Figure 5A). Representative lines of 35S:SlPIF4 (OE#1) and pSlPIF4:SlPIF4 (OE#4) produced 94.0% and 78.7% aborted pollen grains, respectively (Figure 5B). Self-pollination of OE#1 plants did not result in any fruit (Supplemental Figure S7C), whereas crossing OE#1 (♀) × WT (♂) plants resulted in the development of normal fruit and with normal seed numbers (Supplemental Figure S7, C and D), indicating that female fertility was unimpaired in SlPIF4-OE plants.
Figure 5.
Overexpression of SlPIF4 results in pollen infertility by postponing tapetal PCD. A, Overexpression of SlPIF4 leads to pollen sterility. 35S, cauliflower mosaic virus (CaMV) 35S promoter. pSlPIF4, the native SlPIF4 promoter. Each black dot indicates the value of pollen viability for each individual transgenic line. At least six anthers at the mature pollen stage were examined for each plant. Each error bar represents the mean ± se, n = 13 independent 35S:SlPIF4 lines and 14 independent pSlPIF4:SlPIF4 lines. B, Representative images showing pollen viability in WT, 35S:SlPIF4 and pSlPIF4:SlPIF4 lines based on the Alexander method. Ten anthers at the mature pollen stage were examined for each line. C, Tapetal morphology in WT and 35S:SlPIF4 (OE#1) anthers at the early and late uninucleate and early binucleate stages. At least six anthers at different stages were examined for each line. DT, degenerated tapetum; Ml, middle layer; Mt, mitochondrion; Pl, plastid; Pm, plasma membrane; T, tapetum; Ub, Ubisch body; V, vacuole. Bars = 2 µm. D, Transverse section of WT and OE#1 microspores at the early uninucleate (EU) and late uninucleate (LU) stages. DMsp, degenerated microspore; Ex, exine; In, intine; Msp, microspore; V, vacuole. Bars = 2 µm. E, TUNEL assays of tapetal PCD in WT and OE#1 anthers at the early and late uninucleate, and binucleate stages. At least six anthers at different stages were examined for each line. The magenta signal is propidium iodide staining, and the green fluorescence indicates TUNEL positive signal (white arrows). T, tapetum. Bars = 50 µm. F to G, Venn diagram (F) and KEGG analysis (G) of DEGs between WT and OE (35S:SlPIF4) anthers. Numbers in (F) and (G) indicate the number of genes. II, anthers at the tetrad stage; IV, anthers at the late uninucleate stage. H, Expression of DEGs involved in tapetum and pollen wall development in WT and OE (35S:SlPIF4) anthers. Values represent gene expression levels (FPKM) based on RNA-Seq data with three biological replicates. **P <0.01 (DESeq R package). II, anthers at the tetrad stage; IV, anthers at the late uninucleate stage.
To further study tapetal development in the SlPIF4-OE lines, we compared transverse semi-thin sections of developing WT and SlPIF4-OE tapetum from the tetrad to the mature pollen stages (Figure 5C;Supplemental Figure S8A). The tapetum of WT anthers began to degrade at the early uninucleate stage and shrank to a thin and disrupted layer at the early binucleate stage (Figure 5C;Supplemental Figure S8A). In contrast, the SlPIF4- OE tapetum remained intact at the early uninucleate stage and was densely stained at the early binucleate stage (Figure 5C;Supplemental Figure S8A), suggesting that tapetal cell degeneration was delayed in the SlPIF4-OE lines. We also observed a significantly increased number of Ubisch bodies and plastids in the tapetum of SlPIF4-OE anthers at the late uninucleate and early binucleate stages compared to the WT (Figure 5C). Additionally, from the early uninucleate stage onward, the pollen grain walls, which consist of exine and intine, were abnormally thickened, and the microspore cytoplasm and nucleus in SlPIF4-OE lines showed evidence of degradation (Figure 5D;Supplemental Figure S9, A–C). In TUNEL assays, the positive signals in WT anthers began to appear at the tetrad stage, intensified during the early and late uninucleate stages, and disappeared during the early binucleate stage (Figure 5E;Supplemental Figure S8B). However, in SlPIF4-OE anthers, no positive signal was detected until the late uninucleate stage, while strong signals were observed at the early binucleate stage (Figure 5E;Supplemental Figure S8B). These observations indicate that overexpression of SlPIF4 delayed tapetal PCD, which resulted in defects in pollen wall deposition and pollen grain maturation.
To detect transcriptome changes caused by SlPIF4 overexpression in anthers, we performed RNA-Seq analysis of SlPIF4-OE and WT anthers at the tetrad (II) and late uninucleate (IV) stages (Supplemental Figure S10, A and B). A total of 685 DEGs were identified in stage II anthers between SlPIF4-OE and WT plants, with 270 upregulated and 415 downregulated DEGs in SlPIF4-OE anthers (Figure 5F;Supplemental Data Set S4). In contrast, 1,649 DEGs were identified in Stage IV anthers, with 807 upregulated and 842 downregulated DEGs in SlPIF4-OE anthers (Figure 5F). KEGG pathway analysis of DEGs in Stage IV anthers revealed that genes involved in starch/sucrose metabolism, flavonoid biosynthesis, and phenylpropanoid biosynthesis were significantly enriched (Figure 5G).
We measured sugar levels and observed that fructose and glucose contents were significantly lower in Stage IV anthers of the SlPIF4-OE plants compared with WT anthers (Supplemental Figure S10C). The expression levels of SlTDF1, SlDYT1, SlAMS, and SlMYB80 were substantially higher in SlPIF4-OE anthers than in WT, particularly at Stage IV (Figure 5H). Moreover, multiple downstream genes of the transcriptional DYT1-TDF1-AMS-MYB80 cascade, including CYP86C3, SlSKS18, and SlATA1, were significantly upregulated in SlPIF4-OE anthers, while two genes, SlSHT and SlATA7, were downregulated (Figure 5H;Supplemental Table S1). The expression patterns of a subset of these genes were further confirmed by qRT-PCR (Supplemental Figure S11). These results suggest that SlPIF4 affects tapetal PCD by regulating the gene regulatory network during tapetum development and that the over-accumulation of SlPIF4 transcripts suppresses tapetal PCD and pollen development.
The SlPIF4–SlDYT1 heterodimer activates SlTDF1 expression, particularly at MLT
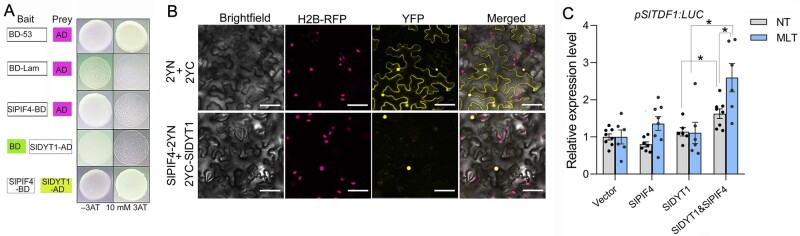
bHLH transcription factors usually function as homodimers or heterodimers (Cui et al., 2016). To investigate whether SlPIF4 physically interacts with SlDYT1, we performed yeast two-hybrid (Y2H) analysis. Yeast strains co-expressing the SlPIF4-binding domain (BD) and activation domain (AD) or SlDYT1-AD and BD did not grow on stringent selection medium (10 mM 3-amino-1,2,4-triazole). However, yeast strains containing SlPIF4-BD and SlDYT1-AD were able to grow on the same selection medium, indicating that SlPIF4 and SlDYT1 interact (Figure 6A). Bimolecular fluorescence complementation (BiFC) assay further confirmed the interaction between SlPIF4 and SlDYT1 in Nicotiana benthamiana containing H2B-RFP (red fluorescent protein) as a nuclear marker (Figure 6B). The localization of SlDYT1 in the nucleus provided a physical basis for its interaction with SlPIF4 (Supplemental Figure S2). The yellow fluorescent protein (YFP) signals in the BiFC assays indicated that the interaction between SlPIF4 and SlDYT1 was strong and specifically occurred in the nucleus (Figure 6B).
Figure 6.
SlPIF4 is required for SlDYT1-mediated transcriptional activation of SlTDF1 under moderately low temperatures. A, Interaction of SlPIF4 with SlDYT1 in Y2H assays. Protein–protein interactions were assessed on SD/-Ade/-His/-Trp/-Leu (SD/-4) medium containing 10 mM 3-amino-1,2,4-triazole (AT). Empty pGBKT7-53 (BD-53) and pGADT7-T1 (AD) vectors were included as the positive control. Empty pGBKT7-Lam and pGADT7-T1 vectors were included as the negative control. B, BiFC confocal images showing SlPIF4 and SlDYT1 interaction in vivo. Nuclear YFP fluorescence was detected in N. benthamiana leaves expressing SlPIF4 fused to the YFP N-terminal region (2YN) and SlDYT1 fused to the YFP C-terminal region (2YC). Empty 2YN and 2YC vectors were included as the positive control. The positions of nuclei are shown by the H2B-RFP marker. Bars = 50 µm. C, SlPIF4 enhances SlDYT1-mediated transcriptional activation of SlTDF1 under moderately low temperature conditions. The LUC/REN mean value of the empty vector pSlTDF1-LUC was set to 1 as the calibrator. Vector, empty vector pSlTDF1-LUC; NT, normal temperature (25°C/20°C, day/night for 3 d); MLT (25°C/20°C, day/night for 2 d, followed by 10°C for 24 h). Each error bar represents the mean ± se, n = 6–8 biological replicates. Asterisks indicate significant differences: **P < 0.05 (Student’s t test).
We then performed a dual-luciferase (LUC) assay to determine the combined effects of SlPIF4-SlDYT1 in activating the SlTDF1 promoter in N. benthamiana leaves grown under NT and MLT conditions (Figure 6C). Following either temperature treatment (NT or MLT), co-transfection of pSlTDF1:LUC with either SlPIF4 or SlDYT1 had no effect, or resulted in only a minor increase, in LUC activity (Figure 6C). However, co-transfection of pSlTDF1:LUC with both SlPIF4 and SlDYT1 resulted in a significant increase in LUC activity driven by the SlTDF1 promoter, and much higher activity was detected following MLT treatment (Figure 6C). These findings suggest that the SlPIF4–SlDYT1 protein complex activates SlTDF1 expression and that this activation can be facilitated by exposure to low temperatures.
SlPIF4 directly regulates SlDYT1 expression
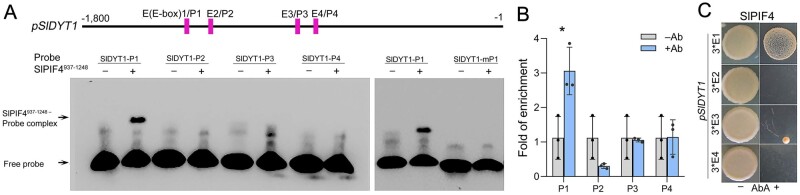
bHLH transcription factors such as DYT1 and AMS regulate gene expression by binding to E-boxes (CANNTG) in the promoters of their target genes (Xu et al., 2010). Given that SlDYT1 was activated in SlPIF4-OE lines, we performed an electrophoretic mobility shift assay (EMSA) to determine whether SlPIF4 directly regulates SlDYT1. A total of four candidate E-box sites (E1–E4) were identified in the 1.8-kb promoter and 5′-untranslated region (UTR) of SlDYT1. EMSA revealed that SlPIF4937–1248-His (a truncated SlPIF4 protein containing the bHLH domain involved in DNA binding and protein oligomerization) fusion protein bound directly to the biotin-labeled E-box-containing probe 1 (P1) of the SlDYT1 promoter, leading to a mobility shift (Figure 7A). However, mutation of the core sequence of the E-box motif in P1 (mP1) abolished this binding (Figure 7A). The binding of SlPIF4 to E-box 1 of the SlDYT1 promoter was further confirmed by quantitative chromatin immunoprecipitation-PCR and yeast one-hybrid (Y1H) assays (Figure 7, B and C). Taken together, these results suggest that SlPIF4 regulates the expression of SlDYT1 by directly binding to E-box1 of its promoter.
Figure 7.
SlPIF4 directly regulates the expression of SlDYT1. A, EMSA showing the binding of the recombinant SlPIF4 protein to the promoter fragment of SlDYT1. DNA oligomers containing an E-box identified in the promoter region of SlDYT1 were labeled with digoxin and used as probes (P). mP1: mutant P1. SlPIF4937–1248, a truncated SlPIF4 protein containing the bHLH domain involved in DNA binding and protein oligomerization. B, Enrichment of the SlDYT1 promoter regions confirmed by ChIP-qPCR with the indicated primer sets (P1, P2, P3, and P4). Each error bar represents the mean ± sd, n = three biological replicates. Asterisk indicates a significant difference at P < 0.05 (Student’s t test). C, Y1H assay showing the binding of SlPIF4 protein to the promoter fragment of SlDYT1. The full-length coding sequence of SlPIF4 was fused to the GAL4 AD in the pGADT7 vector. 3*E: three tandem copies of E-box from the SlDYT1 promoter cloned into the pAbAi vector. Interaction was determined on SD/-Leu medium with AbA (-Leu+AbA300ng/ml). pSlDYT1, the SlDYT1 promoter.
Discussion
SlPIF4 regulates male sterility induced by moderate temperature stress in tomato
PIF4 functions as a hub that integrates temperature signals to control plant morphogenesis and growth (Lucyshyn and Wigge, 2009; Leivar and Monte, 2014; Casal and Balasubramanian, 2019; Dong et al., 2020). However, most studies of PIF4 have been limited to vegetative organs (Xu, 2018), and whether PIF4 is involved in temperature-induced adaptation of plant reproductive organs remains unclear. Plant reproductive development is the phase of the plant lifecycle that is most sensitive to short episodes of extreme temperatures, especially during male gametogenesis (Zinn et al., 2010). However, the underlying regulatory mechanism remains poorly understood. Thus, it is of great importance to study the role of PIF4 in regulating the adaptation of reproductive organs to temperature stress and to determine whether the emerging knowledge of PIF4 in Arabidopsis could be transferred to relevant crop systems.
As PIF4 is a phytochrome-interacting factor, PIF4 is expressed in a circadian rhythm and regulates plant photoperiodic growth (Jeong et al., 2014). Here, we found that SlPIF4 showed a diurnal expression profile in tomato leaves (Figure 1A), which is consistent with a previous report (Rosado et al., 2019). However, SlPIF4 was expressed in a photoperiod-independent manner in tomato flowers (Figure 1A), suggesting that SlPIF4 might have different modes of action or functions in vegetative versus reproductive organs. The expression data pointed to another role for SlPIF4 in anther development, as SlPIF4 transcripts primarily accumulated in microspore mother cells, tapetal cells, and tetrads during early anther development (Figures 1, B and C and 4, C).
SlPIF4-knockout mutants (slpif4) exhibited similar degrees of male fertility to WT plants under NT conditions (Supplemental Figure S3). However, knocking out SlPIF4 increased the tolerance of anthers to moderate temperature stress, especially MLT stress, as indicated by the significant increase in pollen viability and higher fruit set in the slpif4 mutant (Figure 2). In addition, both 35S:SlPIF4 and pSlPIF4:SlPIF4 transgenic lines showed a high percentage of pollen abortion (Figure 5A), suggesting that SlPIF4 is directly involved in regulating pollen development in tomato. Chen et al. (2020) reported that overexpression of GhPIF4 causes pollen abortion in cotton (Gossypium hirsutum). In contrast, in A. thaliana, no pollen development defects were reported in PIF4-OE and pif4 mutant lines (Huq and Quail, 2002; Kumar et al., 2012; Oh et al., 2012; Gangappa et al., 2017). In addition, SlPIF4 shares low protein sequence similarity with AtPIF4 and AtPIF5 (Supplemental Figure S1). Taken together, these results indicate that PIF4 plays different roles in regulating pollen development in A. thaliana and tomato. In summary, our analyses suggest that SlPIF4-mediated male fertility is associated with temperature conditions in tomato plants.
SlPIF4 modulates cold tolerance in anthers by interacting with the tapetum regulatory module
Temperature stress-induced pollen abortion results from tapetum dysfunction at the microspore stage (De Storme and Geelen, 2014; Arshad et al., 2017; Rieu et al., 2017; Feng et al., 2018; Liu et al., 2018; Kiran et al., 2019; Santiago and Sharkey, 2019; Lohani et al., 2020). The correct timing of tapetal PCD and disintegration is indispensable for proper microspore development and pollen maturation and fertility. Upon exposure to low temperatures, tapetal cells display irregular hypertrophy and vacuolation due to delayed PCD, resulting in infertile pollen (De Storme and Geelen, 2014; Arshad et al., 2017; Kiran et al., 2019). In this study, we found that tapetal PCD was significantly delayed in WT anthers after MLT treatment (Figure 3), which is consistent with previous reports (De Storme and Geelen, 2014; Arshad et al., 2017; Kiran et al., 2019). Interestingly, in slpif4 anthers, the MLT-induced delay of tapetal PCD was less pronounced (Figure 3). These results suggest that SlPIF4 affects MLT-mediated male sterility by altering the timing of tapetum PCD. This hypothesis is further supported by the tapetum development phenotypes of SlPIF4-OE anthers, which included delayed tapetal PCD and aberrantly thickened pollen walls (Figure 5, C–E; Supplemental Figures S8 and S9).
Over the past decade, many transcription factors involved in tapetum development and degradation have been identified in A. thaliana, including DYT1, TDF1, AMS, MYB103/MYB80, and bHLH010/bHLH089/bHLH091 (Zhang et al., 2006; Zhu et al., 2008; Ferguson et al., 2017). The genetic DYT1-TDF1-AMS-MYB80 pathway is thought to regulate tapetum development and tapetal PCD in Arabidopsis, and this pathway is also functionally conserved in rice, tomato, and maize (Jung et al., 2005; Li et al., 2006; Niu et al., 2013; Ko et al., 2014; Fu et al., 2014; Jeong et al., 2014; Li et al., 2017). In this transcriptional cascade, DYT1 plays a critical role in regulating early tapetal development by directly activating the expression of its downstream gene TDF1 (Gu et al., 2014). Increased DYT1 expression promotes the accumulation of multiple downstream targets (Ferguson et al., 2017). AMS and MYB80 act downstream of TDF1 and function as master regulators of pollen wall development by directly regulating the expression of their downstream genes involved in callose degeneration (QRT3 and A6), sporopollenin biosynthesis and metabolism (CYP86C3, ACOS5, and SHT), lipid transport (LACS6 and WBC27), and pollen coat formation (EXL4–EXL6; Xu et al., 2010, 2014; Shi et al., 2015). However, knowledge regarding the molecular mechanisms of temperature perception and tolerance in the tapetum remains limited.
Intriguingly, our RNA-Seq data showed that the core transcription factors that regulate tapetal PCD (SlDYT1, SlTDF1, SlAMS, and SlMYB80) and their multiple downstream targets (e.g. SlA6, CYP86C3, SlSKS18, and SlSHT) were responsive to MLT in WT anthers (Figure 4E;Supplemental Figure S5D). The precise control of the expression levels of tapetum-specific transcription factor genes is essential to ensure timely tapetal PCD and pollen development (Sorensen et al., 2003; Ko et al., 2017; Ranjan et al., 2017). We surmise that the induced expression of tapetal-specific transcription factor genes by MLT results in delayed tapetal PCD and pollen abortion in WT anthers. In slpif4 anthers, SlDYT1 expression was induced by MLT to the same level as in WT anthers, while SlTDF1 expression was not induced (Figure 4E), indicating that SlPIF4 is required for SlDYT1-induced activation of SlTDF1 under MLT conditions. Furthermore, the expression of downstream genes regulated by SlTDF1 was less responsive to MLT (Figure 4E;Supplemental Figure S5D), which is consistent with our observation that MLT-induced delayed tapetal PCD was alleviated in slpif4 anthers (Figure 3). In addition, SlPIF4 mRNA and protein levels were significantly higher in WT tapetum and anthers following MLT treatment, and SlPIF4 exhibited a similar expression pattern to SlDYT1 during early anther development (Figure 4F), further indicating that SlPIF4 is a component of the regulatory network controlling tapetum development. In support of this idea, we observed increased expression of these tapetum-specific transcription factor genes and their downstream genes in SlPIF4-OE anthers (Figure 5H;Supplemental Table S1). Together, these findings suggest that SlPIF4 regulates the cold tolerance of anthers by affecting tapetal PCD.
Multiple feed-forward and feed-back loops have been identified in the tapetum transcriptional cascade or among other tapetum-expressed transcription factors to ensure proper protein dynamics and tapetum degeneration (Ferguson et al., 2017). For example, the bHLH transcription factor DYT1 interacts with itself and downstream genes, such as AMS (encoding a bHLH transcription factor), bHLH010, bHLH089, and bHLH091 to regulate anther development (Ferguson et al., 2017). The interactions between DYT1 and bHLH010/bHLH089/bHLH091 promote the nuclear localization of DYT1 and anther development via the activation of TDF1, even though bHLH010/bHLH089/bHLH091 have different functions from DYT1 (Feng et al., 2012; Gu et al., 2014; Zhu et al., 2015; Cui et al, 2016; Ferguson et al., 2017). DYT1 and AMS also modulate their downstream targets by competitively binding to these bHLHs or other tapetum-expressed transcription factors to optimize anther development (Xu et al., 2010; Ferguson et al., 2017). Moreover, bHLH transcription factors can form homo- or heterodimers that bind to specific DNA motifs, and this heterodimerization can affect their DNA binding affinity (Toledo-ortiz et al., 2003; Liu et al., 2013).
In the current study, we demonstrated that SlPIF4 interacts with SlDYT1 (Figure 6, A and B). Importantly, the dual-LUC assay demonstrated that SlDYT1 alone only slightly activated SlTDF1 following either NT or MLT treatment, whereas co-overexpression of SlDYT1 and SlPIF4 resulted in significantly higher activation activity toward SlTDF1, especially under low-temperature conditions (Figure 6C). In addition, SlTDF1 expression was significantly induced by MLT in WT anthers, but not in slpif4 anthers (Figure 4E). These results indicate that the SlDYT1-SlPIF4 heterodimer is required to activate the expression of SlTDF1 under MLT conditions. Notably, Gu et al. (2014) reported that overexpression of TDF1 did not affect fertility in Arabidopsis. However, Gu et al. (2014) expressed AtTDF1 cDNA driven by proDYT1 in dyt1/dyt1 and dyt1/+ plants. Overexpression of AtTDF1 only restored the expression of its downstream genes including AMS, MYB80, MS1, and TEK to WT levels.
Members of the bHLH family bind to the core E-box consensus sequences of their target genes to regulate different biological processes (Carretero-Paulet et al., 2010), and the competitive binding of shared downstream genes to fine-tune growth, development, and defense responses is a common mechanism in both plants and animals (Liao et al., 2007; Hou et al., 2010; Lee et al., 2015). In this study, we demonstrated that SlPIF4 binds specifically to the E-box-containing SlDYT1 promoter (Figure 7) and that SlDYT1 was significantly upregulated in SlPIF4-OE anthers (Figure 5H). These results suggest that SlPIF4 competitively binds to SlDYT1 with its native upstream regulators to fine-tune the tapetum development transcriptional cascade during tomato anther development. Taken together, we propose that SlPIF4 modulates cold tolerance in anthers by interacting with the tapetum regulatory module.
In addition, endogenous JA contents were significantly higher in slpif4 anthers compared to WT anthers under MLT conditions (Supplemental Figure S6). In somatic tissues, JA functions as a positive regulator of cold and freezing tolerance (Hu et al., 2013, 2017; Yang et al., 2019). However, the role of JA in regulating anther development in response to cold stress remains poorly understood. The expression levels of SlDYT1, SlTDF1, and SlMYB80 and endogenous JA contents significantly increased in WT anthers under MLT conditions (Figure 4E;Supplemental Figure S6). However, the expression of SlTDF1 was not induced by MLT in slpif4 anthers, which had significantly higher JA contents than WT anthers under MLT conditions (Figure 4E;Supplemental Figure S6), suggesting that JA does not participate in regulating the transcriptional DYT1-TDF1-AMS-MYB80 cascade in tomato anthers under MLT conditions.
SlPIF4 differentially regulates the cold response during vegetative and reproductive development
PIF4 regulates freezing tolerance in Arabidopsis and tomato seedlings by directly modulating the CBF pathway (Catalá et al., 2011; Lee and Thomashow, 2012; Jiang et al., 2017; Wang et al., 2020). Wang et al. (2020) reported that SlPIF4 positively regulates cold tolerance in tomato seedlings by directly activating CBF1. However, in the current study, knocking out SlPIF4 enhanced the tolerance of tomato anthers to moderate temperature stress (Figure 2), suggesting that SlPIF4 differentially regulates vegetative and reproductive development in response to cold stress. Multiple studies have reported that vegetative and reproductive growth display different levels of temperature sensitivity (Zinn et al., 2010; Lohani et al., 2020). Even at the reproductive growth phase, male organs are generally more sensitive to temperature stress than female organs (Lohani et al., 2020). Furthermore, pollen meiosis I appears to be the most sensitive stage to abiotic stress during the male reproductive phase (Rieu et al., 2017). These findings suggest that the temperature-tolerance mechanisms of vegetative and reproductive organs are different and spatio-temporal-dependent in plants.
To further confirm this notion, we analyzed the expression profiles of CBF genes in both a vegetative organ (leaf) and a reproductive organ (anther) at the tetrad stage in WT and slpif4 plants with and without MLT treatment. Based on qRT-PCR data (Supplemental Figure S12), the transcript levels of both SlCBF1 and SlCBF3 (SlCBF2 was undetectable by qRT-PCR) were significantly reduced in slpif4 leaves under NT conditions (Supplemental Figure S12A), which is consistent with the recent finding that SlPIF4 positively regulates the CBF pathway (Wang et al., 2020). Interestingly, both SlCBF1 and SlCBF3 were significantly upregulated in slpif4 anthers at the tetrad stage (Supplemental Figure S12B). Furthermore, under MLT conditions, both SlCBF1 and SlCBF3 transcripts markedly accumulated in WT anthers but not in slpif4 anthers (Supplemental Figure S12B). These results suggest that the SlPIF4-mediated CBF cold response pathway is not the primary cold acclimation signaling pathway in tomato anthers and that the cold response pathways of vegetative and reproductive organs of tomato plants could be different. In future studies, it would be critical to examine the role of SlPIF4 using tissue-specific promoters.
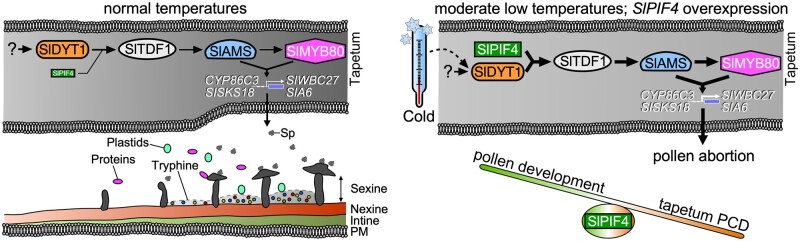
We propose a model in which SlPIF4 operates together with tapetal-specific transcription factors to regulate pollen development in response to MLT in tomato anthers (Figure 8). Under an optimal temperature regime, SlDYT1 directly regulates SlTDF1 during tapetum development and degradation to provide the developing microspores with enzymes, carbohydrates, and nutrients. Under MLT conditions, SlPIF4 enhances SlDYT1-mediated activation of SlTDF1 by forming a SlDYT1–SlPIF4 heterodimer complex with SlDYT1. Overaccumulation of SlTDF1 and its downstream genes postpones tapetal PCD and results in pollen abortion, whereas knocking out SlPIF4 enhances cold tolerance by blocking SlDYT1-mediated activation of SlTDF1 in tomato anthers under MLT conditions. Conversely, overaccumulation of SlPIF4 alters tapetum development by directly activating SlDYT1 or interacting with SlDYT1 to confer male sterility. In summary, our findings reveal that SlPIF4 contributes to the cold response in tomato anthers by modulating the tapetum development-signaling pathway, and they shed light on the molecular mechanism underlying the adaptation of anthers to cold stress.
Figure 8.
Proposed model of the role of SlPIF4 in regulating tapetal and pollen development under normal and moderately low temperatures. Under an optimal temperature regime, SlDYT1 enhances the DYT1-TDF1-AMS-MYB80 transcriptional cascade by directly activating SlTDF1 during tapetum development. Programmed tapetum degradation triggered by the DYT1-TDF1-AMS-MYB80 transcriptional module allows the tapetum to provide the developing microspores with enzymes, carbohydrates, and nutrients. Under moderately low-temperature conditions, SlPIF4 enhances SlDYT1-mediated activation of SlTDF1 by forming a SlDYT1–SlPIF4 heterodimer complex with SlDYT1. Overaccumulation of SlTDF1 and its downstream genes postpones tapetal PCD and further results in pollen abortion. Knocking out SlPIF4 blocks the overactivation of SlTDF1 in tomato anthers at low temperature. While the overaccumulation of SlPIF4 might alter programmed tapetum development by competitively binding to SlDYT1 or by interacting with SlDYT1 under optimal temperature regime. PM, plasma membrane; Sp, sporopollenin precursors.
Materials and methods
Plant materials and growth conditions
Tomato (Solanum lycopersicum) cultivar “Micro-Tom,” provided by the Tomato Genetics Resource Center (University of California, Davis), was used for all gene-transfer experiments and WT controls. WT, slpif4, and SlPIF4-OE plants were grown in a growth chamber under 16-h light with cool-white fluorescent lamps (450–660 nm) at 300 μmol photons m−2 s−1 at 25°C and 8-h dark at 20°C with a relative humidity of 60%–70%. For MLT treatment, tomato plants were grown at 15°C under a 16-h light/8-h dark photoperiod for 6 d. For MHT treatment, tomato plants were grown under a 32°C, 16-h light/27°C, 8-h dark photoperiod for 4 d. Six-week-old tomato plants were used for both MLT and MHT treatment. For Micro-Tom, 6-week-old plants are at the early bloom phase. At this phase, anthers at all six stages (I, early meiosis stage; II, tetrad stage; III, early uninucleate stage; IV, late uninucleate stage; V, binucleate stage; VI, mature pollen stage) appear in the plants, which provided the necessary materials to study anther development at different stages. It is worth noting that tomato anthers at the tetrad stage take approximately 6–7 d to reach the mature pollen stage under an optimal temperature regime. However, in our preliminary assays, we observed that MLT delayed anther development and flowering, while MHT shortened the anther development period and promoted flowering. Under MHT treatment, anthers at the tetrad stage only needed approximately 4–5 d to reach the mature pollen stage. Thus, we selected 4 d as the treatment period of MHT in this study.
Generation of transgenic plants
To develop SlPIF4-expressing transgenic plants, the full-length coding sequence of SlPIF4 was cloned into SmaI and HindIII sites of pCAMBIA1301-3HA vectors (Pan et al., 2016). The pCAMBIA1301-3HA vectors containing the CaMV 35S promoter or the native SlPIF4 promoter with a length of 2,207 bp (−2207 to −1) were used to generate the 35S:SlPIF4-3HA and pSlPIF4:SlPIF4-3HA vectors, respectively. All binary vectors were transformed into Agrobacterium tumefaciens strain GV3101 for transformation into “Micro-Tom” tomato using the leaf-disc method (Sun et al., 2006). The presence of the transgene was confirmed using β-glucuronidase (GUS) staining (Chen et al., 2016) and PCR method. The SlPIF4-knockout mutants (slpif4) were produced using the CRISPR/Cas9 system (Pan et al., 2016), in which a single-guide RNA (sgRNA) was designed to target the bHLH domain of SlPIF4. Two transgene-free homozygous mutants in the T2 generation, slpif4-13 and slpif4-9, containing a 1- and a 6-bp deletion, respectively, were tested in this study. All primer sequences used in this study are listed in Supplemental Data Set S5.
Determination of pollen viability and fruit traits
To determine pollen viability of anthers exposed to temperature stress, anthers at the tetrad stage from the 6-week-old plants were randomly marked with strings before temperature treatments, since the tetrad stage is the most sensitive stage to ambient temperatures (Begcy et al., 2019). We examined pollen viability from the marked anthers at the mature pollen stage after exposure to temperature stress. Pollen viability was determined by staining with fluorescein diacetate or Alexander solution (Alexander, 1969; Wang et al., 2008). Viable and nonviable pollen grains were counted in five different fields under the microscope for each anther or pollen sac. To determine fruit setting rate, 8–10 anthers at the tetrad stage from 6-week-old tomato seedlings were randomly marked by strings for each plant before temperature treatments. A total of 12 individual WT or slpif4 plants were exposed to each temperature treatment (NT, MLT, and MHT). During the breaker stage of fruits, the fruit setting rates were determined for each examined plant. Red fruits diameter and fresh weight were measured with a vernier caliper and digital scale, respectively.
Cytology and microscopy analyses
Tomato flower development stages are closely correlated to flower bud size (Brukhin et al., 2003). In this study, anthers at phases I–IV were assessed and sampled based on flower bud size and morphological characteristics, including the enclosed position of sepals and petals and the color of sepals (Chen et al., 2018). To observe the sexine and nexine layers, pollen grains from anthers at the mature pollen stage were stained with DiOC2 (5 μL mL−1) for 5 min and Tinapol (10 μL mL−1; Sigma, St Louis, MO, USA) for 15 min in the dark (Gu et al., 2014; Lou et al., 2014). Scanning Electron Microscopy and Transmission Electron Microscopy were performed as described previously (Chen et al., 2018). For the TUNEL assays, 10-μm paraffin sections of anthers were prepared and used with a TUNEL apoptosis detection kit (Fluorescein, Roche, Basel, Switzerland) according to the supplier’s instructions. The TUNEL signals were observed under a Nikon confocal laser-scanning microscope (A1-SHS; Nikon, Tokyo, Japan).
Total RNA isolation, qRT-PCR analysis
To investigate the diurnal expression patterns of SlPIF4, leaves and flower buds at the uninucleate stage were sampled with a 4-h interval for 48 h from 6-week-old “Micro-Tom” seedlings that were grown under a 16-h light (300 μmol photons m−2 s−1) at 25°C and 8-h dark at 20°C photoperiod. Total RNA was extracted from the samples using a Total RNA Kit II (OMEGA, Norcross, GA, USA) and reverse-transcribed into cDNA using a PrimeScript RT reagent kit (Takara, Dalian, Liaoning, China). qRT-PCR was performed using SYBR Green Realtime PCR Master Mix (Toyobo, Osaka, Japan) and a Bio-Rad CFX96 Real Time PCR System (Bio-Rad, Hercules, CA, USA) in triplicate for each sample (Pan et al., 2017). The fold changes of gene expression were calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001). Briefly, the fold changes of targeted gene expression were expressed relative to the internal control gene SlUBI3. Ct data were produced by qRT-PCR, and, where sample 0 represents root or anthers at the tetrad stage or leaf in the corresponding figures.
In situ hybridization
Anthers at different developmental stages (I–IV) were collected from WT tomato plants, fixed, dehydrated, embedded, sectioned, and attached to Superfrost Plus Microscope Slides (Thermo Scientific, Waltham, MA, USA), as previously described (Chen et al., 2018). A 291-bp SlPIF4 cDNA fragment was amplified and used as the template for transcription with SP6 and T7 RNA polymerases to produce digoxigenin-labeled RNA probes (Roche, Basel, Switzerland). As a negative control, sense RNA probes were synthesized and hybridized to sections of tomato flower buds. The in situ hybridization signals were observed under a Nikon microscope (ECLIPSE 90i; Nikon, Tokyo, Japan).
Subcellular localization
The full-length coding sequences of SlPIF4 and SlDYT1 were inserted into the BamH I and Xba I sites of pFGC-eGFP vector (Chen et al., 2018) to generate the SlPIF4-eGFP and SlDYT1-eGFP fusion genes driven by a CaMV35S promoter. Agrobacterium tumefaciens cultures containing the SlPIF4-eGFP, SlDYT1-eGFP, or pFGC-eGFP construct were independently infiltrated into 4-week-old N. benthamiana leaves expressing the red nuclear marker RFP-H2B (Chen et al., 2018). After 48 h of incubation, fluorescent signals were observed under a Nikon confocal laser scanning microscope (A1-SHS; Nikon, Tokyo, Japan).
Transcriptome profiling
Anthers at the early uninucleate stage (III) were collected from slpif4 and WT plants grown under MLT or NT conditions. Anthers at the tetrad (II) and late uninucleate stage (IV) were collected from SlPIF4-OE (35S:SlPIF4) and WT plants grown under NT conditions. All samples were collected in three biological replicates, each containing at least 20 anthers. Total RNA was isolated from the samples using the TRIZOL method (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. DNA was removed using a DNA-free kit (Ambion, Austin, TX, USA). RNA-Seq libraries were prepared using a NEBNext Ultra RNA Library Prep Kit (NEB, Ipswich, MA, USA) following the manufacturer’s recommendations and sequenced by Novogene Biotech (Beijing, China) on the HiSeq 4000 platform (Illumina, Inc., San Diego, CA, USA). Raw reads were processed to remove low quality and adapter sequences and those with >10% of “N” base. The cleaned reads were aligned to the tomato reference genome SL3.0 (The Tomato Genome Consortium, 2012) using Hisat2 v2.0.5 (Kim et al., 2019). FeatureCounts v1.5.0-p3 (Liao et al., 2014) was used to count the number of reads mapped to each gene, and the resulting raw counts were normalized to Fragments Per Kilobase of transcript per Million mapped fragments (FPKM). DEGs were identified using the DESeq R package (Anders and Huber, 2010) with an adjusted P < 0.05 and fold change ≥2. The expression levels of selected DEGs were confirmed by qRT-PCR. Gene ontology (GO) enrichment and KEGG analyses were performed using the Plant MetGenMAP (Joung et al., 2009) and KEGG database (Kanehisa and Goto, 2000), respectively.
Immunoblotting
Equal amounts of WT anthers at stages II–III were sampled at 0, 24, 48, 72, and 96 h after MLT treatment. To extract proteins, the anther samples were ground in liquid nitrogen and resuspended in extraction buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 5 mM DTT, and 5% Triton) with freshly added 1 mM phenylmethylsulfonyl fluoride and 1× protease inhibitor cocktail (Sigma, St Louis, MO, USA). Protein concentrations were determined by Coomassie staining (Bradford, 1976). Protein samples were fractionated by SDS–PAGE (sodium dodecyl sulphate–polyacrylamide gel electrophoresis) and electro-transferred to a nitrocellulose membrane (GE Healthcare, Germany). Immunoblot analysis was performed using antibodies against PIF4 (Agrisera, Vännäs, Västerbotten, Sweden; Wang et al., 2020) and plant actin (ABclonal, Wuhan, Hubei, China).
Yeast one-hybrid assay
Yeast one-hybrid assays were carried out using the Matchmatch Gold Yeast One-Hybrid System (Clontech, Mountain View, CA, USA). Three tandem copies of DNA fragments, each containing an E-box site from the SlDYT1 promoter, were cloned into the pAbAi vector, and the full-length coding sequence of SlPIF4 was fused to the GAL4 AD in the pGADT7 vector. The pAbAi baits were transformed into Y1HGold cells, which were screened on selective dropout (SD)-Ura medium with different concentrations of aureobasidin A (AbA) to determine the minimal inhibitory concentration. The AD-prey vector was then transformed into the bait strain and screened on SD/-Leu/AbA medium.
Yeast two-hybrid assay
Y2H assays were performed using the Clontech Matchmaker GAL4 System (Clontech, Mountain View, CA, USA) according to the manufacturer’s instructions. The full-length coding sequences of SlPIF4 and SlDYT1 were cloned into the pGBKT7 and pGADT7 vectors to produce the SlPIF4-BD and SlDYT1-AD constructs, respectively. Bait constructs were transformed into yeast strain AH109 and tested for auto-activation. Yeast strains containing both the bait and prey constructs were screened on SD/-Trp-Leu agar medium and SD/-His-Ade-Trp-Leu medium with 10 mM 3-amino-1,2,4 triazole (3 AT) to confirm positive interactions.
BiFC assay
The full-length coding sequences of SlPIF4 and SlDYT1 were cloned into the N-terminus of YFP in the p2YN vector and the C-terminus of YFP in the p2YC vector at PacI and AscI sites (Chen et al., 2018) to produce the SlPIF4-2YN and 2YC-SlDYT1 constructs, respectively. The constructs were individually transformed into A. tumefaciens strain C58C1 cells, which were incubated and resuspended in infiltration buffer (10 mM MES, 10 mM MgCl2, and 10 μM acetosyringone) to the same concentration with the OD 600 (optical densities at 600 nm) = 1.0. Equal volumes of A. tumefaciens cultures were immediately mixed and infiltrated into N. benthamiana leaves using a needleless syringe (Chen et al., 2018). After 48 h of incubation at 25°C, the YFP fluorescent signals were detected under a Nikon confocal laser scanning microscope (A1-SHS; Nikon, Tokyo, Japan).
Electrophoretic mobility shift assay
The pET-32a-His-SlPIF4937–1248 vector was generated using a truncated sequence of SlPIF4 containing the bHLH domain. His-tagged SlPIF4 protein was expressed in Escherichia coli strain Rosetta (DE3) and purified using a High Affinity Ni-NTA Resin kit (GenScript, Piscataway, NJ, USA). DNA fragments of the SlDYT1 promoter containing the E-box and mutated E-box region were synthesized as biotin-labeled oligonucleotides. EMSA was performed using a LightShift Chemiluminescent EMSA Kit (Thermo Fisher Scientific, Gaithersburg, MD, USA).
ChIP-qPCR assay
ChIP-quantitative PCR (qPCR) assays were performed using an EpiQuik Plant ChIP Kit following the manufacturer’s protocol (Epigentek, Farmingdale, NY, USA). Briefly, equal amounts of anthers at the tetrad and uninucleate stages (a total of 1 g) sampled from 35S:SlPIF4-3HA transgenic plants were cross-linked in 1% formaldehyde to produce chromatin complexes, which were immunoprecipitated with an anti-HA antibody (Abcam, Cambridge, MA, USA). Goat anti-mouse IgG (Millipore, Rockville, MD, USA) was used as the negative control. qPCR analysis was used to detect the enrichment of the chromatin fragments. SlACTIN2 was used as an internal reference.
Dual-LUC assays
The full-length cDNA of SlPIF4 or SlDYT1 was cloned into the SacI and KpnI sites of pGreen II 0029 62-SK vector, and the SlTDF1 promoter was inserted into the KpnI and NcoI sites of pGreen II 0800-LUC vector (Hellens et al., 2005). All constructs were individually transformed into A. tumefaciens strain GV3101. A mixture of cells containing constructs with protein and promoter sequences was infiltrated into N. benthamiana according to a previous protocol (Zhang et al., 2018). The activity of the transcription factor binding to the promoter was measured on the third day post-infiltration, as indicated by the ratio of enzyme activities of LUC and REN (Renilla) using a Dual Luciferase Assay Kit (Promega, Madison, WI, USA). For NT treatment, after infiltration, N. benthamiana seedlings were grown in a growth chamber under a cycle of 16-h light (∼150 μmol photons m−2 s−1, cool-white fluorescent lamps at 450–660 nm) at 25°C and 8-h dark at 20°C for 3 d. For MLT treatment, the seedlings were grown in a growth chamber under a cycle of 16-h light at 25°C and 8-h dark at 20°C for 2 d after infiltration, then were transferred to low temperature (10°C) conditions for 24 h.
HPLC analysis of endogenous sugar contents
To measure endogenous sugar contents in SlPIF4-OE and WT plants, anthers at the late uninucleate stage were collected. Fresh anthers (200 mg) were ground to a powder in liquid nitrogen and homogenized in 5 mL of 80% ethanol. The samples were incubated at 80°C for 30 min and centrifuged at 12,000g for 20 min at room temperature. The supernatant was collected, dried by evaporation, and resuspended in 3 mL distilled deionized water. After filtration (0.22 μm nylon membrane), the supernatant was analyzed by HPLC (Agilent1100, Santa Clara, CA, USA) using a Waters Spherisorb NH2 column (4.6 mm × 250 mm, 5 μm) at 35°C. The mobile phase was 80% acetonitrile at a flow rate of 1 mL/min for 22 min. The injection volume was 10 μL, while sucrose, D-(−)-fructose, and D-(+)-glucose (Sigma‐Aldrich, St. Louis, MO, USA) of different gradient concentrations were used as standards. Six independent biological replicates were performed at the same time.
Determination of endogenous IAA, JA, and ABA levels
To measure endogenous IAA, JA, and ABA contents, pooled samples composed of at least 20 anthers at the tetrad stage were collected from 15 individual plants exposed to NT or MLT for 6 d for each biological replicate. Three independent biological replicates were performed for each treatment. Measurement of endogenous hormones was performed as previously reported (Fu et al. 2012) with minor modifications (Pan et al. 2019). Briefly, fresh anthers were sampled and ground into a powder in liquid nitrogen using a mortar and pestle. For each sample, 100 mg powder was homogenized in 1 mL of ethyl acetate that was spiked with d2‐IAA (Sigma‐Aldrich, St. Louis, MO, USA), d5-JA (QCC), and d6-ABA (OlchemIm Ltd, Olomouc, Czech Republic) with agitation for 10 min. The supernatant was collected after centrifugation at 13,000g for 20 min at 4°C and dried with N2 gas. The residue was resuspended in 0.2 mL of 60% (v/v) methanol and centrifuged at 13,000g for 10 min at 4°C. The supernatant was filtered through a 0.22-μm nylon membrane and analyzed by HPLC (high-performance liquid chromatography) /MS (mass spectrometry) –MS on an Agilent 1290 infinity HPLC system coupled with an Agilent 6460 Triple Quad LC/MS device (Agilent Technologies, Santa Clara, CA, USA). An Agilent Zorbax XDB C18 column (150 mm × 2.1 mm, 3.5 μm) was used to perform HPLC analysis. Three independent biological replicates were performed for each sample.
Statistical analysis
Student’s t test and ANOVA results are provided in Supplemental Data Set S6.
Accession numbers
Sequence data from this article can be found in the Sol Genomics Database (https://solgenomics.net/) under the following accession numbers: SlPIF4 (Solyc07g043580), SlDYT1 (Solyc02g079810), SlTDF1 (Solyc03g059200), SlAMS (Solyc08g062780), SlMYB80 (Solyc10g005760), SlCYP86C3 (Solyc02g014730), SlSKS18 (Solyc04g081520), SlATA1 (Solyc11g018600), SlSHT (Solyc07g015960), SlATA7 (Solyc07g053780), SlOleosin (Solyc06g069260), SlVGD1 (Solyc06g084620), SlCBF1 (Solyc03g026280), and SlCBF3 (Solyc03g026270). The RNA-seq data are available from the NCBI Sequence Read Archive under accession code PRJNA631095 (https://www.ncbi.nlm.nih.gov/sra/PRJNA631095).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of SlPIF4, AtPIF4, and AtPIF5 full-length protein sequences.
Supplemental Figure S2. Subcellular localization of SlPIF4 and SlDYT1 in N. benthamiana.
Supplemental Figure S3. Analysis of pollen viability in SlPIF4-knockout mutants (slpif4) and WT plants under normal conditions.
Supplemental Figure S4. Knocking out SlPIF4 enhances tolerance of tomato pollen to moderately low-temperature stress.
Supplemental Figure S5. Analysis of RNA-Seq data from WT and slpif4 mutants under NT and MLT conditions.
Supplemental Figure S6. Endogenous IAA, JA, and ABA contents in WT and slpif4 anthers at the tetrad stage under NT and MLT conditions.
Supplemental Figure S7. Flower and fruit development in WT and SlPIF4-overexpressing lines.
Supplemental Figure S8. Tapetal morphology and PCD in WT and SlPIF4-overexpressing (OE#1) lines.
Supplemental Figure S9. Pollen exine and intine development in WT and SlPIF4-overexpressing (OE#1) lines.
Supplemental Figure S10. RNA-Seq analysis and sugar contents in WT and SlPIF4-overexpressing lines.
Supplemental Figure S11. Expression analysis of SlPIF4, SlDYT1, SlTDF1, SlAMS, and SlMYB80 in WT and SlPIF4-overexpressing (OE, 35S:SlPIF4) anthers by qRT-PCR.
Supplemental Figure S12. Expression analysis of SlCBF1 and SlCBF3 in WT and slpif4 plants under NT and MLT conditions.
Supplemental Table S1. DEGs involved in pollen and pollen wall development in WT and SlPIF4-overexpressing (OE, 35S:SlPIF4) anthers.
Supplemental Data Set S1. Transcriptome analysis of WT anthers grown under NT and MLT conditions.
Supplemental Data Set S2. Transcriptome analysis of slpif4 anthers grown under NT and MLT conditions.
Supplemental Data Set S3. Transcriptome analysis of slpif4 and WT anthers grown under NT or MLT conditions.
Supplemental Data Set S4. Transcriptome analysis of SlPIF4-overexpressing and WTanthers.
Supplemental Data Set S5. Primer sequences used in this study.
Supplemental Data Set S6. Statistical analysis data.
Supplementary Material
Acknowledgments
We thank Jingquan Yu, Qiaomei Wang, and Yanhong Zhou (Zhejiang University, China) for technical support of the sugar analysis and immunoblotting assay and Xinai Zhao (Heidelberg University, German) for support and advice during the project.
Funding
This work was supported by grants from the National Key Research and Development Program of China (2018YFD1000800), the National Natural Science Foundation of China (31772316 and 31471878), the Natural Science Foundation of the Zhejiang Province, China (LZ17C150002), and the United States National Science Foundation (IOS-1855585).
Conflict of interest statement. None declared.
These authors contributed equally to this work (C.P., D.Y.).
G.L., Z.F., and C.P. conceived the study. C.P., D.Y., L.Y., M.L., and F.Y. generated transgenic lines and performed cytological observation and yeast one/two-hybrid assays; D.Y. and Y.L. carried out the protein analysis; C.P., X.Z., and Z.F. performed the data analysis, C.P., X.Z., D.Y., A.T.L., and S.M.A. performed the temperature stress experiments. C.P., G.L., and Z.F. wrote and revised the article. All authors discussed and commented on the final manuscript.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) are: Gang Lu (glu@zju.edu.cn) and Zhangjun Fei (zf25@cornell.edu).
References
- Alexander MP (1969) Differential staining of aborted and nonaborted pollen. Stain Technol 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T, Toriyama K (2011) Genetic regulation of sporopollenin synthesis and pollen exine development. Annu Rev Plant Biol 62: 437–460 [DOI] [PubMed] [Google Scholar]
- Arshad MS, Farooq M, Asch F, Krishna JSV, Prasad PVV, Siddique KHM (2017) Thermal stress impacts reproductive development and grain yield in rice. Plant Physiol Biochem 115: 57–72 [DOI] [PubMed] [Google Scholar]
- Baron KN, Schroeder DF, Stasolla C (2012) Transcriptional response of abscisic acid (ABA) metabolism and transport to cold and heat stress applied at the reproductive stage of development in Arabidopsis thaliana. Plant Sci 188–189: 48–59 [DOI] [PubMed] [Google Scholar]
- Begcy K, Nosenko T, Zhou LZ, Fragner L, Weckwerth W, Dresselhaus T (2019) Male sterility in maize after transient heat stress during the tetrad stage of pollen development. Plant Physiol 181: 683–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brukhin V, Hernould M, Gonzalez N, Chevalier C, Mouras A (2003) Flower development schedule in tomato Lycopersicon esculentum cv sweet cherry. Sex Plant Reprod 15: 311–320 [Google Scholar]
- Carretero-Paulet L, Galstyan A, Roig-Villanova I, Martínez-García JF, Bilbao-Castro JR, Robertson DL (2010) Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol 153: 1398–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon A, Shen H, Huq E (2007) Phytochrome interacting factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci 12: 514–521 [DOI] [PubMed] [Google Scholar]
- Casal JJ, Balasubramanian S (2019) Thermomorphogenesis. Annu Rev Plant Biol 70:321–346 [DOI] [PubMed] [Google Scholar]
- Catalá R, Medina J, Salinas J (2011) Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proc Natl Acad Sci U S A 108: 16475–16480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Guan X, Qin L, Zou T, Zhang Y, Wang J, Wang Y, Pan C, Lu G (2016) Downregulation of the mitogen-activated protein kinase SlMAPK7 gene results in pollen abortion in tomato. Plant Cell Tissue Organ Cult 126: 79–92 [Google Scholar]
- Chen L, Yang D, Zhang Y, Wu L, Zhang Y, Ye L, Pan C, He Y, Huang L, Ruan YL. et al. (2018) Evidence for a specific and critical role of mitogen-activated protein kinase 20 in uni-to-binucleate transition of microgametogenesis in tomato. New Phytol 219: 176–194 [DOI] [PubMed] [Google Scholar]
- Chen M, Xie S, Wang CZ, Li YL, Zhang XL, Min L (2020) Mechanism of GhPIF4 regulating anther abortion under high temperature stress in cotton. Acta Agronom Sin 46: 1368–1379 [Google Scholar]
- Cui J, You C, Zhu E, Huang Q, Ma H, Chang F (2016) Feedback regulation of DYT1 by interactions with downstream bHLH factors promotes DYT1 nuclear localization and anther development. Plant Cell 28: 1078–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Storme N, Geelen D (2014) The impact of environmental stress on male reproductive development in plants: biological processes and molecular mechanisms. Plant Cell Environ 37: 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Yan Y, Jiang B, Shi Y, Jia Y, Cheng J, Shi Y, Kang J, Li H, Zhang D, et al. (2020) The cold response regulator CBF1 promotes Arabidopsis hypocotyl growth at ambient temperatures. EMBO J 39: e103630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falasca G, D'Angeli S, Biasi R, Fattorini L, Matteucci M, Canini A, Altamura MM (2013) Tapetum and middle layer control male fertility in Actinidia deliciosa. Ann Bot 112: 1045–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B, Lu D, Ma X, Peng Y, Sun Y, Ning G, Ma H (2012) Regulation of the Arabidopsis anther transcriptome by DYT1 for pollen development. Plant J 72: 612–624 [DOI] [PubMed] [Google Scholar]
- Feng B, Zhang C, Chen T, Zhang X, Tao L, Fu G (2018) Salicylic acid reverses pollen abortion of rice caused by heat stress. BMC Plant Biol 18: 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson AC, Pearce S, Band LR, Yang C, Ferjentsikova I, King J, Yuan Z, Zhang D, Wilson ZA (2017) Biphasic regulation of the transcription factor ABORTED MICROSPORES (AMS) is essential for tapetum and pollen development in Arabidopsis. New Phytol 213: 778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Yu J, Cheng X, Zong X, Xu J, Chen M, Li Z, Zhang D, Liang W (2014) The rice basic helix-loop-helix transcription factor TDR INTERACTING PROTEIN2 is a central switch in early anther development. Plant Cell 26: 1512–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Chu J, Sun X, Wang J, Yan C (2012) Simple, rapid, and simultaneous assay of multiple carboxyl containing phytohormones in wounded tomatoes by UPLC-MS/MS using single SPE purification and isotope dilution. Anal Sci 28: 1081–1087 [DOI] [PubMed] [Google Scholar]
- Gangappa SN, Berriri S, Kumar SV (2017) PIF4 coordinates thermosensory growth and immunity in Arabidopsis. Curr Biol 27: 243–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorno F, Wolters-Arts M, Mariani C, Rieu I (2013) Ensuring reproduction at high temperatures: the heat stress response during anther and pollen development. Plants 2: 489–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu JN, Zhu J, Yu Y, Teng XD, Lou Y, Xu XF, Liu JL, Yang ZN (2014) DYT1 directly regulates the expression of TDF1 for tapetum development and pollen wall formation in Arabidopsis. Plant J 80: 1005–1013 [DOI] [PubMed] [Google Scholar]
- Hatfield JL, Prueger JH (2015) Temperature extremes: Effect on plant growth and development. Weather Clim Extremes 10: 4–10 [Google Scholar]
- Hedhly A, Hormaza JI, Herrero M (2009) Global warming and sexual plant reproduction. Trends Plant Sci 14: 30–36 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Lee LYC., Xia K, Yan Y, Yu H (2010) DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell 19: 884–894 [DOI] [PubMed] [Google Scholar]
- Hu Y, Jiang L, Wang F, Yu D (2013) Jasmonate regulates the inducer of CBF expression–c-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 25: 2907–2924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Jiang Y, Han X, Wang H, Pan J, Yu D (2017) Jasmonate regulates leaf senescence and tolerance to cold stress: crosstalk with other phytohormones. J Exp Bot 68: 1361–1369 [DOI] [PubMed] [Google Scholar]
- Huq E, Quail PH (2002) PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J 21: 2441–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong HJ, Kang JH, Zhao M, Kwon JK, Choi HS, Bae JH, Lee HA, Joung YH, Choi D, Kang BC (2014) Tomato Male sterile 1035 is essential for pollen development and meiosis in anthers. J Exp Bot 65: 6693–6709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang B, Shi Y, Zhang X, Xin X, Qi L, Guo H, Li J, Yang S (2017) PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis. Proc Natl Acad Sci U S A 114: E6695–E6702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung JG, Corbett AM, Fellman SM, Tieman DM, Klee HJ, Giovannoni JJ, Fei Z (2009) Plant MetGenMAP: an integrative analysis system for plant systems biology. Plant Physiol 151: 1758–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KH, Han MJ, Lee YS, Kim YW, Hwang I, Kim MJ, Kim YK, Nahm BH, An G (2005) Rice undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell 17: 2705–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucl Acids Res 28: 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Hwang G, Kim S, Thi TN, Kim H, Jeong J, Kim J, Kim J, Choi G, Oh E (2020) The epidermis coordinates thermoresponsive growth through the phyB-PIF4-auxin pathway. Nat Commun 11: 1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL (2019) Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37: 907–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiran A, Kumar S, Nayyar H, Sharma KD (2019) Low temperature-induced aberrations in male and female reproductive organ development cause flower abortion in chickpea. Plant Cell Environ 42: 2075–2089 [DOI] [PubMed] [Google Scholar]
- Ko SS, Li MJ, Ku MS, Ben Ho YC, Lin YJ, Chuang MH, Hsing HX, Lien YC, Yang HT, Chang HC, et al. (2014) The bHLH142 transcription factor coordinates with TDR1 to modulate the expression of EAT1 and regulate pollen development in rice. Plant Cell 26: 2486–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko SS, Li MJ, Lin YJ, Hsing HX, Yang TT, Chen TK, Jhong CM, Ku MS (2017) Tightly controlled expression of bHLH142 is essential for timely tapetal programmed cell death and pollen development in rice. Front Plant Sci 8: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku S, Yoon H, Suh HS, Chung YY (2003) Male-sterility of thermosensitive genic male-sterile rice is associated with premature programmed cell death of the tapetum. Planta 217: 559–565 [DOI] [PubMed] [Google Scholar]
- Kumar SV, Lucyshyn D, Jaeger KE, Alós E, Alvey E, Harberd NP, Wigge PA (2012) Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484: 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CM, Thomashow MF (2012) Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc Natl Acad Sci U S A 109: 15054–15059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Hnilova M, Maes M, Lin YCL., Putarjunan A, Han SK, Avila J, Torii KU (2015) Competitive binding of antagonistic peptides fine-tunes stomatal patterning. Nature 522: 439–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N, Choi G (2017) Phytochrome-interacting factor from Arabidopsis to liverwort. Curr Opin Plant Biol 35: 54–60 [DOI] [PubMed] [Google Scholar]
- Leivar P, Quail PH (2011) PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E (2014) PIFs: systems integrators in plant development. Plant Cell 26: 56–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DD, Xue JS, Zhu J, Yang ZN (2017) Gene regulatory network for tapetum development in Arabidopsis thaliana. Front Plant Sci 8: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhang D, Liu H, Yin C, Li X, Liang W, Yuan Z, Xu B, Chu H, Wang J (2006) The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18: 2999–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao B, Hu Y, Brewer G (2007) Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat Struct Mol Biol 14: 511–518 [DOI] [PubMed] [Google Scholar]
- Lin L, Liu X, Yin R (2018) PIF3 integrates light and low temperature signaling. Trends Plant Sci 23: 93–95 [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W (2014) FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930 [DOI] [PubMed] [Google Scholar]
- Liu B, Mo WJ, Zhang D, De Storme N, Geelen D (2019) Cold influences male reproductive development in plants: a hazard to fertility, but a window for evolution. Plant Cell Physiol 60: 7–18 [DOI] [PubMed] [Google Scholar]
- Liu L, Fan XD (2013) Tapetum: regulation and role in sporopollenin biosynthesis in Arabidopsis. Plant Mol Biol 83: 165–175 [DOI] [PubMed] [Google Scholar]
- Liu Y, Li X, Li K, Liu H, Lin C (2013) Multiple bHLH proteins form heterodimers to Mediate CRY2-dependent regulation of flowering-time in Arabidopsis. PLoS Genet 9: e1003861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Shi X, Li S, Zhang L, Song X (2018) Oxidative stress and aberrant programmed cell death are associated with pollen abortion in isonuclear alloplasmic male-sterile wheat. Front Plant Sci 9: 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lobell DB, Gourdji SM (2012) The influence of climate change on global crop productivity. Plant Physiol 160: 1686–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohani N, Singh MB, Bhalla PL (2020) High temperature susceptibility of sexual reproduction in crop plants. J Exp Bot 71: 555–568 [DOI] [PubMed] [Google Scholar]
- Lou Y, Xu XF, Zhu J, Gu JN, Blackmore S, Yang ZN (2014) The tapetal AHL family protein TEK determines nexine formation in the pollen wall. Nat Commun 5: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucyshyn D, Wigge PA (2009) Plant development: PIF4 integrates diverse environmental signals. Curr Biol 19: R265–R266 [DOI] [PubMed] [Google Scholar]
- Ma H (2005) Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu Rev Plant Biol 56: 393–434 [DOI] [PubMed] [Google Scholar]
- Min L, Li Y, Hu Q, Zhu L, Gao W, Wu Y, Ding Y, Liu S, Yang X, Zhang X (2014) Sugar and auxin signaling pathways respond to high-temperature stress during anther development as revealed by transcript profiling analysis in cotton. Plant Physiol 164: 1293–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller F, Rieu I (2016) Acclimation to high temperature during pollen development. Plant Reprod 29: 107–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan GL, Zhai J, Arikit S, Morrow D, Fernandes J, Mai L, Nguyen N, Meyers BC, Walbot V (2017) MS23, a master basic helix-loop-helix factor, regulates the specification and development of the tapetum in maize. Development 144: 163–172 [DOI] [PubMed] [Google Scholar]
- Niu N, Liang W, Yang X, Jin W, Wilson ZA, Hu J, Zhang D (2013) EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat Commun 4: 1–11 [DOI] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Wang ZY (2012) Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol 14: 802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omidi M, Siahpoosh MR, Mamghani R, Modarresi M (2014) The influence of terminal heat stress on meiosis abnormalities in pollen mother cells of wheat. Cytologia 79: 49–58 [Google Scholar]
- Oshino T, Abiko M, Saito R, Ichiishi E, Endo M, Kawagishi-Kobayashi M, Higashitani A (2007) Premature progression of anther early developmental programs accompanied by comprehensive alterations in transcription during high-temperature injury in barley plants. Mol Genet Genom 278: 31–42 [DOI] [PubMed] [Google Scholar]
- Paik I, Kathare PK, Kim J Il, Huq E (2017) Expanding roles of PIFs in signal integration from multiple processes. Mol Plant 10: 1035–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C, Ye L, Qin L, Liu X, He Y, Wang J, Chen L, Lu G (2016) CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci Rep 6: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C, Ye L, Zheng Y, Wang Y, Yang D, Liu X, Chen L, Zhang Y, Fei Z, Lu G (2017) Identification and expression profiling of microRNAs involved in the stigma exertion under high-temperature stress in tomato. BMC Genomics 18: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C, Yang D, Zhao X, Jiao C, Yan Y, Lamin-Samu AT, Wang Q, Xu X, Fei Z, Lu G (2019) Tomato stigma exertion induced by high temperature is associated with the jasmonate signalling pathway. Plant Cell Environ 42: 1205–1221 [DOI] [PubMed] [Google Scholar]
- Peng Z, Cheng L, He YJ, Wang J, Guan XY, Liu SY, Lu G (2013) Cytological study on microsporogenesis of Solanum lycopersicum var. Micro-Tom under high temperature stress. Acta Ecol Sin 33: 2084–2092 [Google Scholar]
- Phan HA, Iacuone S, Li SF, Parish RW (2011) The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. Plant Cell 23: 2209–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M, Delker C, Franklin KA, Wigge PA, Halliday KJ, Van Zanten M (2016) Molecular and genetic control of plant thermomorphogenesis. Nat Plants 2: 15190. [DOI] [PubMed] [Google Scholar]
- Ranjan R, Khurana R, Malik N, Badoni S, Parida SK, Kapoor S, Tyagi AK (2017) BHLH142 regulates various metabolic pathway-related genes to affect pollen development and anther dehiscence in rice. Sci Rep 7: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieu I, Twell D, Firon N (2017) Pollen development at high temperature: from acclimation to collapse. Plant Physiol 173: 1967–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado D, Gramegna G, Cruz A, Lira BS, Freschi L, De Setta N, Rossi M (2016) Phytochrome interacting factors (PIFs) in Solanum lycopersicum: diversity, evolutionary history and expression profiling during different developmental processes. PLoS One 11: 1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado D, Trench B, Bianchetti R, Zuccarelli R, Alves FRR, Purgatto E, Floh EIS, Nogueira FTS, Freschi L, Rossi M (2019) Downregulation of phytochrome-interacting factor 4 influences plant development and fruit production. Plant Physiol 181: 1360–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago JP, Sharkey TD (2019) Pollen development at high temperature and role of carbon and nitrogen metabolites. Plant Cell Environ 42: 2759–2775. [DOI] [PubMed] [Google Scholar]
- Sharma KD, Nayyar H (2016) Regulatory networks in pollen development under cold stress. Front Plant Sci 7: 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Cui M, Yang L, Kim YJ, Zhang D (2015) Genetic and biochemical mechanisms of pollen wall development. Trends Plant Sci 20: 741–753 [DOI] [PubMed] [Google Scholar]
- Smith LM (2019) The heat is on: maize pollen development after a heat wave. Plant Physiol 181: 387–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen AM, Kröber S, Unte US, Huijser P, Dekker K, Saedler H (2003) The Arabidopsis aborted microspores (ams) gene encodes a MYC class transcription factor. Plant J 33: 413–423 [DOI] [PubMed] [Google Scholar]
- Sun HJ, Uchii S, Watanabe S, Ezura H (2006) A highly efficient transformation protocol for Micro-Tom, a model cultivar for tomato functional genomics. Plant Cell Physiol 47: 426–431 [DOI] [PubMed] [Google Scholar]
- Thakur P, Kumar S, Malik JA, Berger JD, Nayyar H (2010) Cold stress effects on reproductive development in grain crops: an overview. Environ Exp Bot 67: 429–443 [Google Scholar]
- The Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng C, Zhang H, Hammond R, Huang K, Meyers BC, Walbot V (2020) Dicer-like 5 deficiency confers temperature-sensitive male sterility in maize. Nat Commun 11: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaka D, Nakashima K, Maruyama K, Kidokoro S, Osakabe Y, Ito Y, Matsukura S, Fujita Y, Yoshiwara K, Ohme-Takagi M, et al. (2012) Rice phytochrome-interacting factor-like protein OsPIL1 functions as a key regulator of internode elongation and induces a morphological response to drought stress. Proc Natl Acad Sci U S A 109: 15947–15952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-ortiz G, Huq E, Quail PH (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcay-Barrena G, Wilson ZA (2006) Altered tapetal PCD and pollen wall development in the Arabidopsis ms1 mutant. J Exp Bot 57: 2709–2717 [DOI] [PubMed] [Google Scholar]
- Wang F, Chen X, Dong S, Jiang X, Wang L, Yu J, Zhou Y (2020) Crosstalk of PIF4 and DELLA modulates CBF transcript and hormone homeostasis in cold response in tomato. Plant Biotechnol J 18: 1041–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Schauer N, Usadel B, Frasse P, Zouine M, Hernould M, Latché A, Pech JC, Fernie AR, Bouzayen M (2009) Regulatory features underlying pollination-dependent and -independent tomato fruit set revealed by transcript and primary metabolite profiling. Plant Cell 21: 1428–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang WZ, Song LF, Zou JJ, Su Z, Wu WH (2008) Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol 148: 1201–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong SX, Lu JY, Lou Y, Teng XD, Gu JN, Zhang C, Shi QS, Yang ZN, Zhu J (2016) The transcription factors MS188 and AMS form a complex to activate the expression of CYP703A2 for sporopollenin biosynthesis in Arabidopsis thaliana. Plant J 88: 936–946 [DOI] [PubMed] [Google Scholar]
- Xu D (2018) Multifaceted roles of PIF4 in plants. Trends Plant Sci 23: 749–751 [DOI] [PubMed] [Google Scholar]
- Xu J, Ding Z, Vizcay-Barrena G, Shi J, Liang W, Yuan Z, Werck-Reichhart D, Schreiber L, Wilson ZA, Zhang D (2014) Aborted microspores acts as a master regulator of pollen wall formation in Arabidopsis. Plant Cell 26: 1544–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Yang C, Yuan Z, Zhang D, Gondwe MY, Ding Z, Liang W, Zhang D, Wilson ZA (2010) The aborted microspores regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana. Plant Cell 22: 91–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wang R, Wang Y, Zhang L, Yao S (2020) A point mutation in LTT1 enhances cold tolerance at the booting stage in rice. Plant Cell Environ 43: 992–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashino T, Nomoto Y, Lorrain S, Miyachi M, Ito S, Nakamichi N, Fankhauser C, Mizuno T (2013) Verification at the protein level of the pif4-mediated external coincidence model for the temperature-adaptive photoperiodic control of plant growth in Arabidopsis thaliana. Plant Signal Behav 8: e23390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Duan G, Lin C, Liu G, Han Y (2019) The crosstalks between jasmonic acid and other plant hormone signaling highlight the involvement of jasmonic acid as a core component in plant response to biotic and abiotic stresses. Front Plant Sci 10: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Han J, Kim YJ, Song M, Yang Z, He Y, Fu R, Luo Z, Hu J, Liang W, Zhang D (2017) Two rice receptor-like kinases maintain male fertility under changing temperatures. Proc Natl Acad Sci U S A 114: 12327–12332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Sun Y, Timofejeva L, Chen C, Grossniklaus U, Ma H (2006) Regulation of Arabidopsis tapetum development and function by dysfunctional Tapetum1 (DYT1) encoding a putative bHLH transcription factor. Development 133: 3085–3095 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yin X, Xiao Y, Zhang Z, Li S, Liu X, Zhang B, Yang X, Grierson D, Jiang G, Klee HJ, Chen K (2018) An ethylene response factor-MYB transcription complex regulates furaneol biosynthesis by activating quinone oxidoreductase expression in strawberry. Plant Physiol 178: 189–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu E, You C, Wang S, Cui J, Niu B, Wang Y, Qi J, Ma H, Chang F (2015) The DYT1-interacting proteins bHLH010, bHLH089 and bHLH091 are redundantly required for Arabidopsis anther development and transcriptome. Plant J 83: 976–990 [DOI] [PubMed] [Google Scholar]
- Zhu J, Chen H, Li H, Gao JF, Jiang H, Wang C, Guan YF, Yang ZN (2008) Defective in tapetal development and function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. Plant J 55: 266–277 [DOI] [PubMed] [Google Scholar]
- Zinn KE, Tunc-Ozdemir M, Harper JF (2010) Temperature stress and plant sexual reproduction: Uncovering the weakest links. J Exp Bot 61: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.