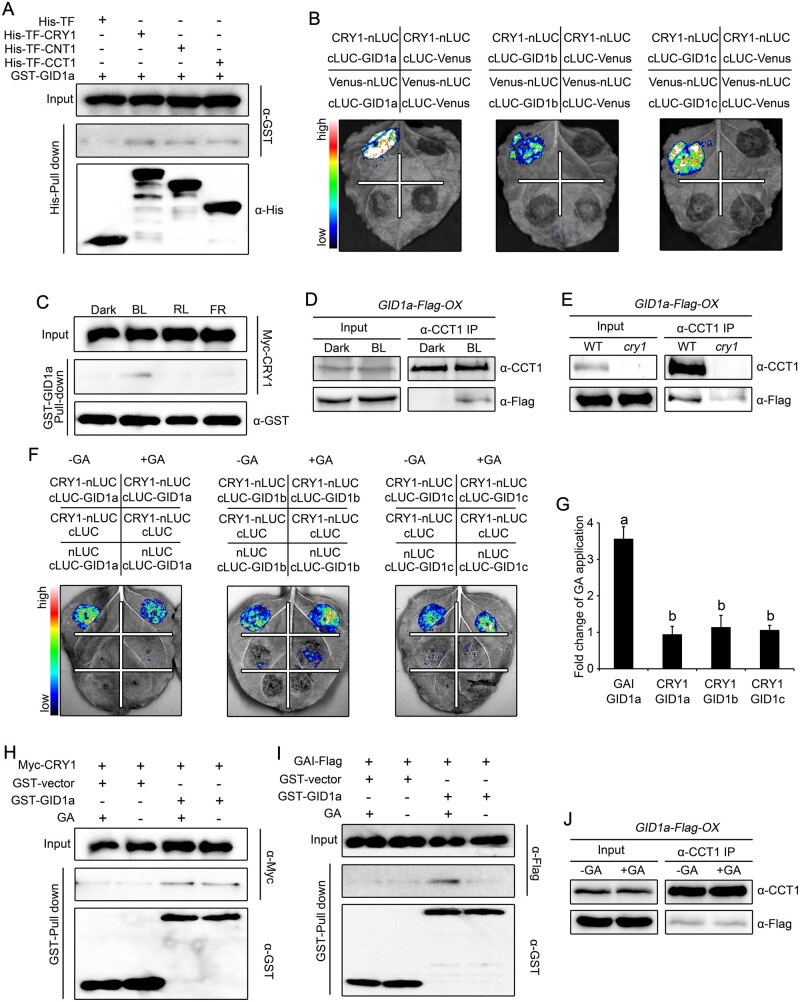
Figure 5.
CRY1 interacts with GID1a in a blue light-dependent but GA-independent manner. A, Pull-down assays showing the interaction of CRY1, CNT1, and CCT1 with GID1a. His-TF-CRY1, His-TF-CNT1, and His-TF-CCT1 served as baits and were detected with anti-His antibody. GST–GID1a served as prey and was detected with anti-GST antibody. Two independent experiments were performed, and one is shown. B, Split-luciferase complementation imaging assays showing the interaction of CRY1 with GID1a, GID1b, and GID1c. Venus-nLUC and cLUC-Venus served as negative controls. Two independent experiments were performed, and one is shown. C, Cell-free GST pull-down assays showing the blue light-specific interaction of CRY1 with GID1a. GST–GID1a served as bait. Preys were protein extracts prepared from dark-adapted Myc-CRY1-OX seedlings that were exposed to blue light (BL, 30 μmol·m–2·s–1), red light (RL, 50 μmol·m–2·s–1) or far-red light (FR, 10 μmol/m2/s) for 1 h. Two independent experiments were performed, and one is shown. D, E, Co-IP assays showing the blue light-dependent interaction of CRY1 with GID1a in Arabidopsis. Dark-adapted GID1a-Flag-OX (D) and cry1 GID1a-Flag-OX (E) seedlings were maintained in the dark or exposed to blue light (50 μmol·m–2·s–1) for 1 h, followed by immunoprecipitation with anti-CCT1 antibody. The IP (CRY1) and co-IP signals (GID1a) were detected in immunoblots probed with anti-CCT1 and anti-Flag antibodies, respectively. Two independent experiments were performed, and one is shown. F, G, Split-luciferase complementation imaging assays illustrating the interaction of CRY1 with GID1 in a GA-independent manner in N. benthamiana cells. Application of 100-μM GA3 did not affect the interaction of CRY1 with GID1a, GID1b, or GID1c (F). The quantification of luciferase activity for the samples in (F) is shown in (G). Data are shown as means of biological triplicates ± sd (n = 4). Letters “a” and “b” indicate statistically significant differences for the indicated values, as determined by a one-way analysis of variance (ANOVA), followed by Tukey’s least significant difference (LSD) test (P < 0.05). H, I, Cell-free GST pull-down assays showing the GA-independent interaction of CRY1 with GID1. GST–GID1a served as bait. Preys were protein extracts prepared from dark-adapted Myc-CRY1-OX (H) and GAI-Flag-OX (I) seedlings exposed to blue light (50 μmol·m–2·s–1) for 1 h. Extracts were mixed with GST–GID1a, and then 100-μM GA3 was added (+) or not (−). Two independent experiments were performed, and one is shown. J, Co-IP assays showing the GA-independent interaction of CRY1 with GID1 in Arabidopsis. Dark-adapted GID1a-Flag-OX seedlings were exposed to blue light (50 μmol·m–2·s–1) for 1 h, and followed by immunoprecipitation with an anti-CCT1 antibody without (–) or with (−) 100-μM GA3 added. The IP (CRY1) and co-IP signals (GID1a) were detected in immunoblots probed with anti-CCT1 and anti-Flag antibodies, respectively. Two independent experiments were performed, and one is shown.