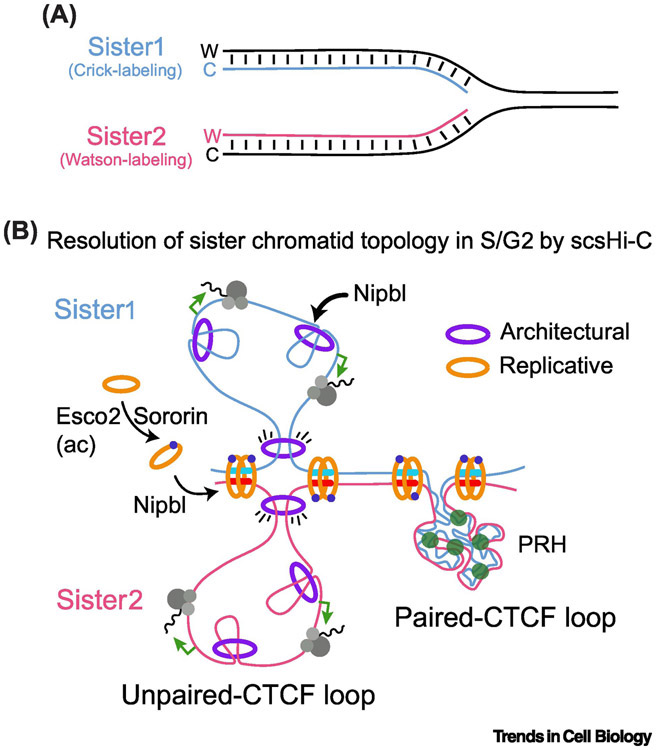
Figure 3. The topology of replicated genome.
(A) By the specific labelling of Watson (W) or Crick(C) strands during DNA replication in S phase, sister-chromatid-sensitive Hi-C approach (scsHi-C) allowed the resolution of chromatin contacts between sister chromatids at S and G2 stages of the cell cycle. (B) Two classes of CTCF loops emerged. Paired CTCF loops, with high level of trans interactions between sister chromatids that correspond to polycomb-repressed heterochromatin (PRH). In contrast, unpaired CTCF loops show abundant cis interactions and poor inter-chromatid connection that correlate with active transcription. In both cases, Sororin and Esco2 stabilizes replicative cohesin showing enrichment at CTCF loop boundaries.