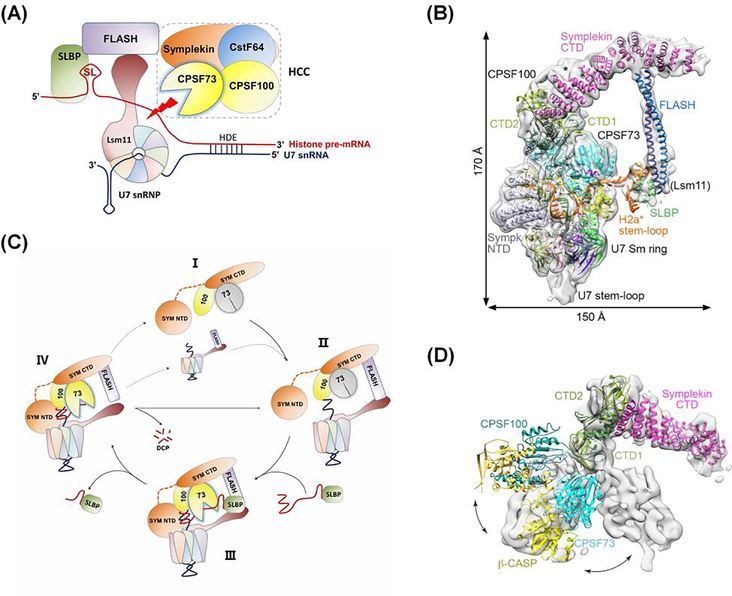
Figure 2. The role of CPSF73 in processing unpolyadenylated histone mRNAs.
(A) The histone mRNA 3’ end processing machinery. The cleavage site is defined by two conserved elements: a stem-loop (SL) upstream of the cleavage site that is bound by stem-loop binding protein (SLBP), and the histone downstream element (HDE) which base-pairs with U7 small nuclear ribonucleoprotein (snRNP) downstream of the cleavage site. The histone cleavage complex (HCC) contains CPSF73, CPSF100, and symplekin, and also CstF64, the subunit of CstF that recognizes the downstream G/U-rich element of polyadenylated mRNA precursors. U7 snRNP interacts with the FLASH protein to recruit HCC to the cleavage site (red arrow). (B) Structure of the histone processing machinery assembled on RNA containing the histone H2A processing site, as determined by cryo-electron microscopy (reproduced from [19]). The positions of the N-Terminal Domain (NTD) and C-terminal Domain (CTD) of symplekin and the two interacting subdomains of the CTDs of CPSF73 and CPSF100 (CTD1 and CTD2) are indicated. CstF64 was not observed in the EM density and is not required for cleavage in vitro. (C) Proposed model for histone pre-mRNA 3’ end processing cycle (adapted from [19]). The U7 snRNP, FLASH, and HCC (State I) come together to form the processing complex (State II), followed by the recognition of the pre-mRNA bound to SLBP, activation of CPSF73 and pre-mRNA cleavage (State III; see also Figure 2B). After cleavage, the upstream product bound to SLBP is released, the downstream product is degraded from the 5’ end (state IV), and the machinery can be recycled.
(D) Conformational differences in the positions of CPSF73, CPSF100, and symplekin relative to each other in the inactive state of mCF (gray surface) compared to that of active HCC (reproduced from [19]).