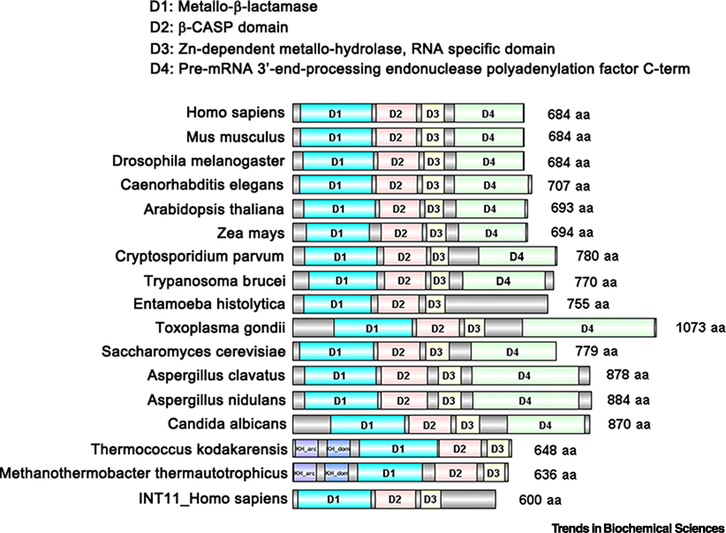
Figure I. Domain organization of CPSF73: comparison from prokaryotes to eukaryotes.
The structural domains and sequence patterns of selected CPSF3 homologs were predicted by InterPro Databases and UniProt Knowledgebase (UniProtKB) using the name of each protein, and were then visually compared with the conserved regions by DOG 2.0 [88]. The Metallo-β-lactamase domain is indicated in blue, the β-CASP domain in red, the Zn-dependent metallo-hydrolase, RNA specificity domain in yellow and the pre-mRNA 3’-end processing endonuclease polyadenylation factor C-term in green. The origin of each protein is at the left; the size in amino acid (aa) residues is at the right. CPSF73 homologs are shown from mammals (Homo sapiens and Mus musculus), other model metazoans (Drosophila melanogaster and C. elegans), plants (Arabidopsis thaliana and Zea mays), parasites (Cryptosporidium parvum, Trypanosoma brucei, Entamoeba histolytica, and Toxoplasma gondii), fungus (Saccharomyces cerevisiae, Aspergillus clavatus, Aspergillus nidulans, and Candida albicans), and Archae bacteria (Thermococcus kodakarensis and Methanothermobacter thermautotrophicus).