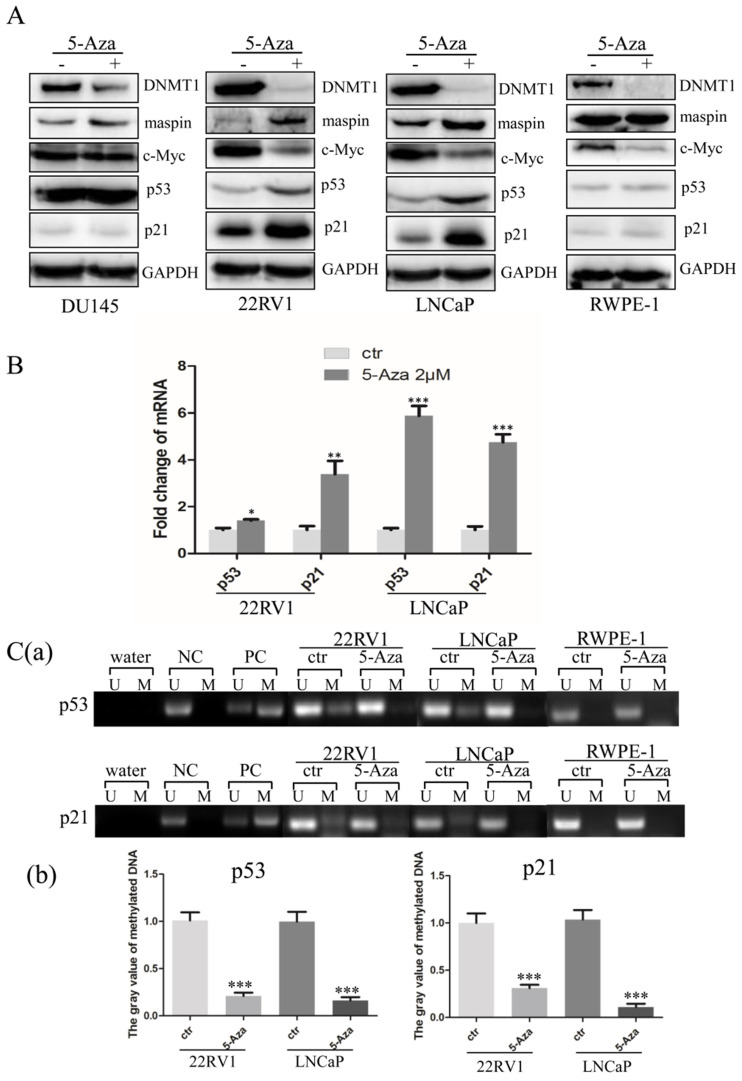
Figure 5.
5-Aza activated p53 and p21 gene transcription and downregulated c-Myc expression in prostate cancer cells. (A) DU145, 22RV1, LNCaP and RWPE-1cells were treated with 5-Aza at 2 μM for 96 h. The cells were harvested and the expression of DNMT1, maspin, c-Myc, p53 and p21 proteins was determined by western blotting. GAPDH was used as an internal loading control. (B) The 22RV1 and LNCaP were treated 5-Aza, as described above. The cells were harvested, the total cellular RNA was extracted, and qRT-PCR assay was performed to determine the mRNA levels of p53 and p21, as described in the Materials and Methods section. The bars denote the mean ± SD, * P<0.05, ** P<0.01, *** P<0.001. (C) 22RV1, LNCaP and RWPE-1 cells were treated with 5-Aza at 2 μM for 96 h. Next, the cellular genomic DNA was treated with sodium bisulfite to convert unmethylated cytosines to uracil. Methylation-specific PCR analysis was performed and the PCR products were separated on non-denaturing polyacrylamide gels. The bands reflecting methylated and unmethylated DNA of p53 (a) and p21 (b) were visualized by staining with ethidium bromide. ImageJ software was used to analyze the gray value of methylated DNA of p53 (c) and p21 (d). The bars denote the mean ± SD, *** P<0.001.Abbreviations: 5-Aza, 5-Aza-2′-deoxycytidine; ctr, control; DNMT1, DNA methyltransferase 1; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; M: methylated; maspin, mammary serine protease inhibitor; NC, unmethylated negative control DNA; PARP, poly (ADP-ribose) polymerase; PC, methylated positive control DNA; qRT-PCR, quantitative reverse transcription-polymerase chain reaction; SD, standard deviation; U: unmethylated; Water, blank control of ddH2O.