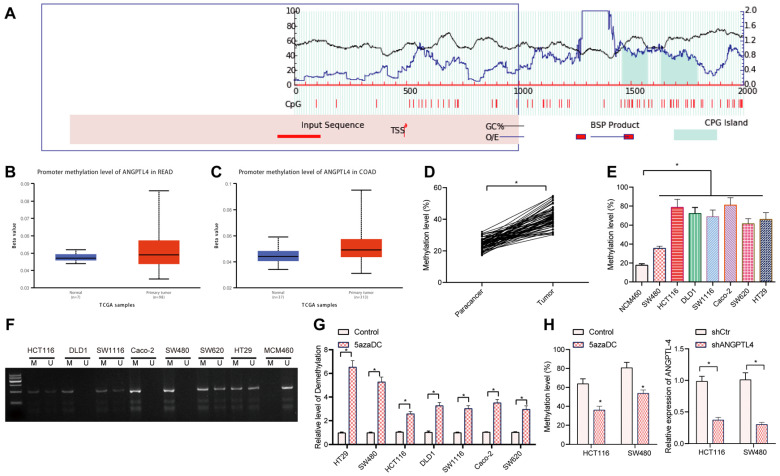
Figure 2.
DNA methylation mediated the expression of ANGPTL4 in CRC. (A) Methylation islands of genes predicted using MethPrimer. (B) Methylation of the promoter region of ANGPTL4 in rectal cancer samples included in the TCGA database; blue boxplots indicate normal samples and red boxplots indicate tumor samples (n = 24, B: p = 1.56280E-04, C: p = 3.40250E-12). (C) Methylation of the promoter region of ANGPTL4 in CRC samples included in the TCGA database; blue boxplots indicate normal samples and red boxplots indicate tumor samples (n = 24, B: p = 1.56280E-04, C: p = 3.40250E-12). (D) Bisulfite sequencing analysis of methylation levels in CRC clinical samples (n = 67, * p < 0.05 vs. para-carcinoma tissues). (E) Bisulfite sequencing was used to determine the methylation level of ANGPTL4 promoter region in CRC cell lines (* p < 0.05 vs. NCM460 cells). (F) Methylation-specific PCR was used to determine the methylation status of each CRC cell line. (G) RT-qPCR was used to determine the expression levels of ANGPTL4 in CRC cell lines after 5-aza-DC treatment (* p < 0.05 vs. the control group). (H) Bisulfite sequencing was used to detect the methylation level of ANGPTL4 promoter region in SW480 and HCT116 cells after 5-aza-DC treatment (* p < 0.05 vs. the control group).