Figure 3. Linking genes to molecules using metabolomics and transcriptomics.
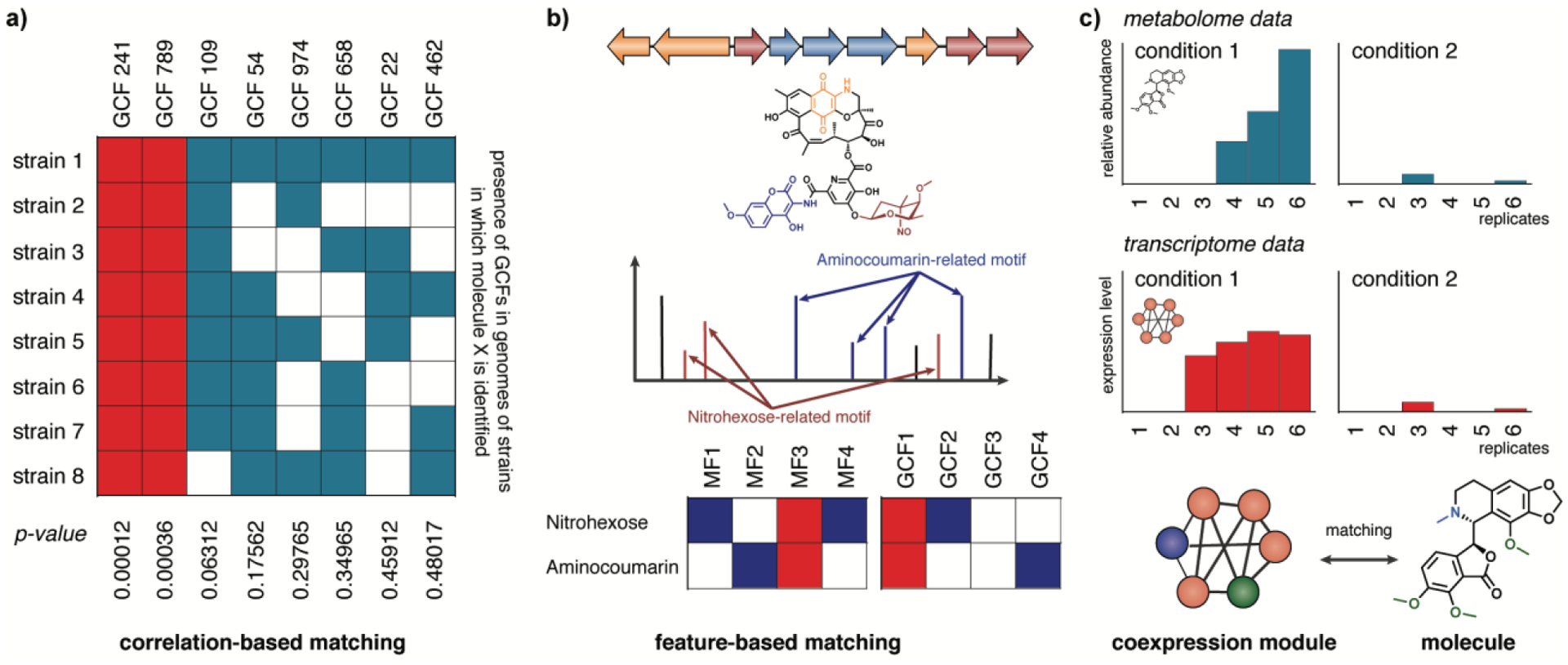
Several approaches have been developed to link metabolites to genes and gene clusters encoding their biosynthesis. a) In bacteria, pattern-based genome mining approaches have been developed that match families of molecules (related by spectral similarity) to gene clusters families (GCFs, related by sequence similarity) through metabologenomic correlation123, which identifies which GCFs co-occur strongly in the same strains where a given metabolite is observed. b) Molecules can also be connected to genes and gene clusters through feature-based matching, in which chemical features (substructures and modifications that are either manually annotated or identified using algorithms that identify motifs in MS/MS data) are linked to genes and gene modules that are known to be responsible for the biosynthesis of such features. c) Transcriptomic data can also be used to identify potential biosynthetic pathways for a molecule of interest by, for example, identifying modules of coexpressed genes whose expression correlates with the presence of a given metabolite across a range of divergent conditions (for example, different biological stresses136).
