Figure 6.
Cebpg deletion results in reduced IFNγ production in NK cells and blocking IFNγ signaling improves HSC fitness
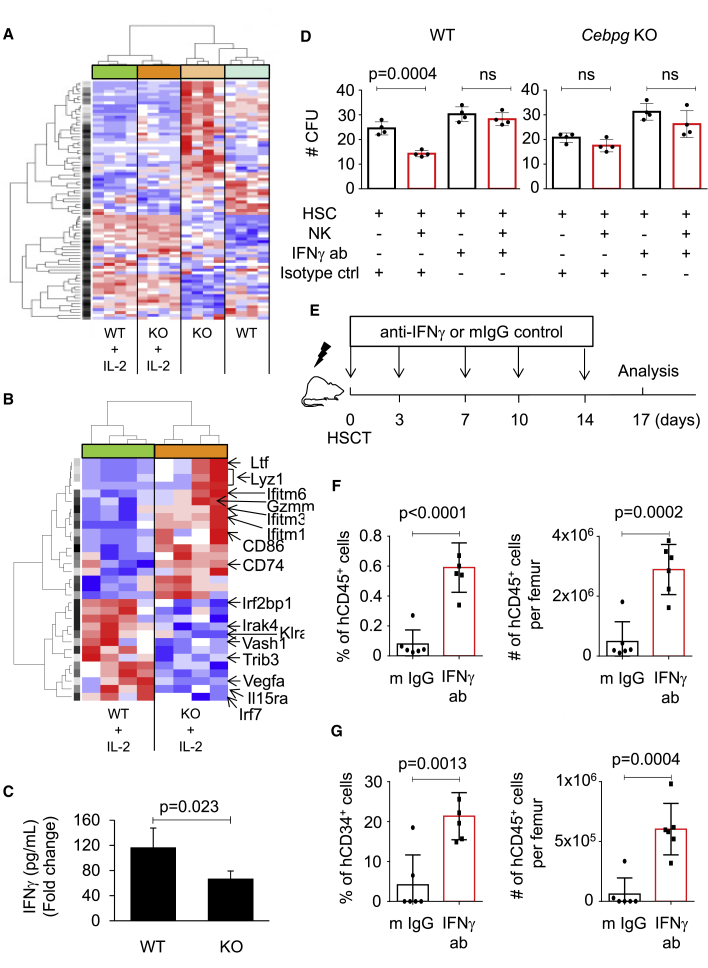
(A) Heatmap and hierarchical clustering based on gene expression of 84 probe sets, which compose the NK signature. NK cells were isolated from WT and Cebpg KO murine SPs in non-stimulation conditions or upon stimulation with the NK cell-activating cytokine IL-2 (n = 4 for each condition). Data were normalized to z scores for each gene. Red/blue color indicates increase/decrease in gene expression relative to the universal mean for each gene. See also Table S1.
(B) Heatmap and hierarchical clustering according to expression of 29 genes present in the NK signature. WT and Cepbg KO NK cells were treated with IL-2 prior to gene expression profile analysis. Several genes deregulated between the two groups are indicated.
(C) IFNγ levels in supernatants after culturing overnight WT or Cebpg KO NK cells. Cultures were established in the absence or presence of IL-2. Y axes indicate cytokine levels (pg/mL) related to the condition without IL-2 for WT and KO NK cells (fold change). n = 4 mice per group, two independent experiments.
(D) Colony culture assays of murine cells. Y axes indicate the number of CFU. X axes indicate culture conditions: NK cells correspond to WT or Cebpg KO mice as indicated. Each dot represents one culture well, two independent experiments were performed.
(E) Illustration of the experimental scheme. Arrows and numbers indicate days when treatment was administered. Seventeen days after transplantation, recipient NSG mice were sacrificed and analyzed.
(F and G) Relative percentage (left graphics) and absolute number of cells per femur (right graphics). Y axes indicate the percentage (%) and numbers (#) of human CD45+ cells (F) and CD34+ cells (G) in BM. Black boxes indicate values for mice that received treatment with mouse IgG control antibodies, and red boxes indicate values for mice that received IFNγ-blocking antibody treatment. Each animal is indicated by a symbol (n = 5–6 animals per group). For (C), (D), (F) and (G), data represent mean ± SD, two-tailed Student's t test was used to assess statistical significance (p values are indicated; ns, not significant).