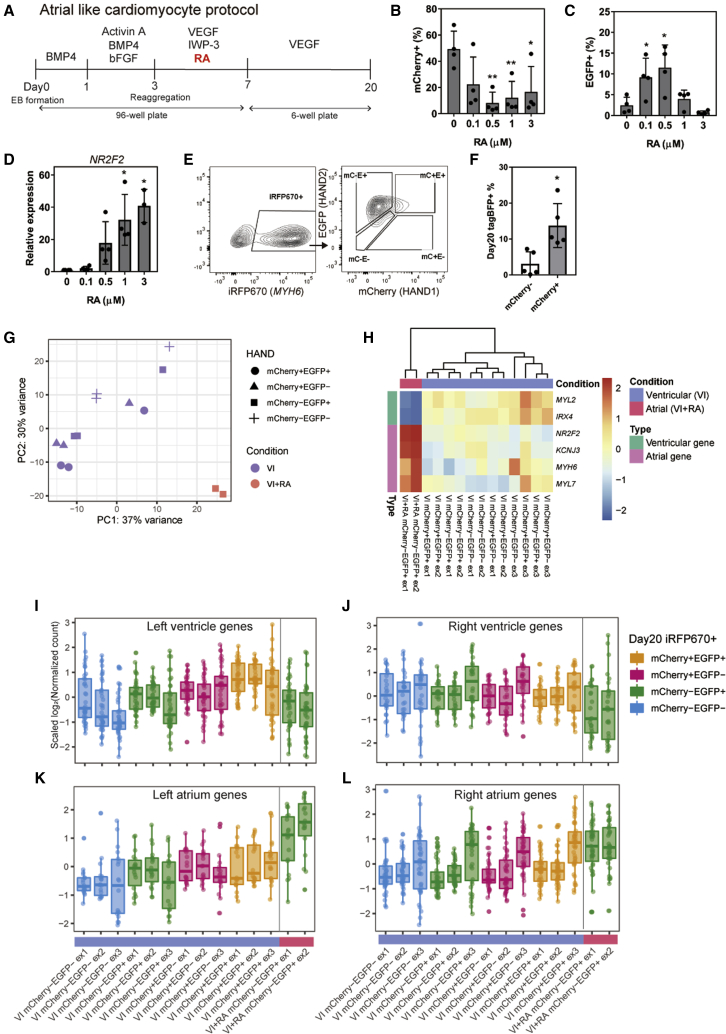
Figure 4.
Retinoic acid in the atrial protocol upregulated HAND2 and downregulated HAND1
(A) Scheme of the RA-modified atrial CM differentiation protocol.
(B and C) Percentages of mCherry+ cells (B) and EGFP+ cells (C) on day 7 at different RA concentrations (n = 4 independent experiments). ∗p < 0.05, ∗∗p < 0.01 by one-way ANOVA with Dunnett's test comparing with 0 μM.
(D) Gene expression level of NR2F2, an atrial marker gene, on day 7 at different RA concentrations. (n = 3–4 independent experiments). ∗p < 0.05, ∗∗p < 0.01 by one-way ANOVA with Dunnett's test comparing with 0 μM.
(E) Representative FACS plots of iRFP670, mCherry (mC), and EGFP (E) expressions on day 20 (n = 5 independent experiments).
(F) Percentages of tagBFP+ cells on day 20. EBs were mixed with tagBFP+ mCherry+ cells and tagBFP− cells or with tagBFP+ mCherry− cells and tagBFP− cells on day 5 (n = 3 independent experiments). ∗p < 0.05 by unpaired t test. Data represent means ± SD.
(G) Principal component analysis plot of ventricular CMs (VEGF + IWP-3 on day 3; VI) and atrial CMs (VEGF + IWP-3 + RA on day 3; VI + RA) (n = 3 independent experiments).
(H) Heatmap of scaled expression levels of MYL7, MYH6, KCNJ3, and NR2F2 (atrial marker genes) and MYL2 and IRX4 (ventricular marker genes) from the RNA-seq data of day 20 CMs.
(I–L) Box plots of the scaled log2 normalized counts of chamber-specific genes in each subpopulation. The upper and lower quartiles are indicated by the boxes, and the median by the lines within each box.