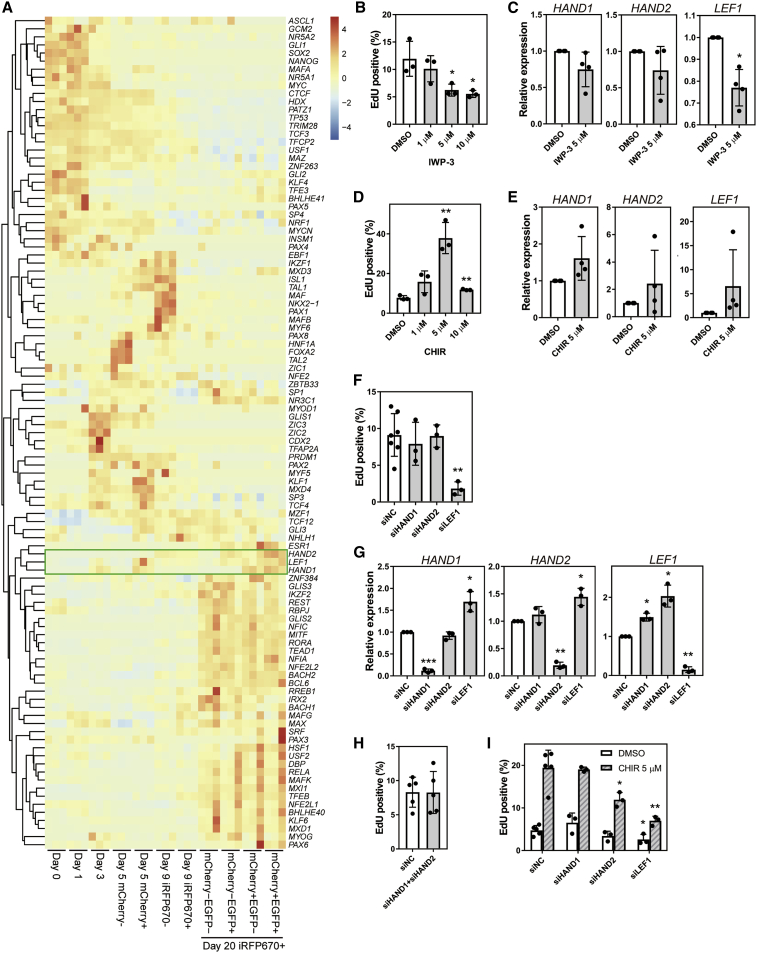
Figure 6.
WNT signaling and proliferation in CMs
(A) Heatmap and clustering of scaled expression levels of 103 TFs predicted as upstream factors from the RNA-seq data of days 0, 3, 5 (isolated mCherry− and mCherry+ populations), day 9 (isolated iRFP670− and iRFP670+ populations), and day 20 subpopulations in iRFP670+ CMs (n = 3 independent experiments). Green box highlights HAND1, LEF1, and HAND2.
(B) Percentage of EdU+ cells on day 20 among EGFP+ CMs isolated on day 15 following WNT inhibitor (IWP-3) administration on days 16–18 (n = 3 independent experiments). ∗p < 0.05 by one-way ANOVA with Dunnett's test comparing with DMSO.
(C) Real-time qPCR results for HAND1, HAND2, and LEF1 expression under IWP-3 treatment (n = 4 independent experiments). ∗p < 0.05, by Welch's t test.
(D) Percentage of EdU+ cells on day 20 among EGFP+ CMs isolated on day 15 following GSK3 inhibitor CHIR99021 (CHIR) administration on days 16–18 (n = 3 independent experiments). ∗∗∗p < 0.001 by one-way ANOVA with Dunnett's test comparing with DMSO.
(E) Real-time qPCR results of HAND1, HAND2, and LEF1 expression under CHIR treatment (n = 4 independent experiments).
(F) Percentage of EdU+ cells after treatment with siRNAs for negative control (siNC), HAND1 (siHAND1), HAND2 (siHAND2), and LEF1 (siLEF1) (n = 3–7 independent experiments). ∗∗p < 0.01 by one-way ANOVA with Dunnett's test comparing with siNC.
(G) Real-time qPCR results for HAND1, HAND2, and LEF1 expression after siRNA treatment on day 20. EGFP+ CMs isolated on day 15 (n = 3 independent experiments). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 by Welch's t test.
(H) Percentage of EdU+ cells with simultaneous knockdown of HAND1 and HAND2 (n = 5 independent experiments).
(I) Percentage of EdU+ cells after CHIR and siRNA combination treatment (n = 3–5 independent experiments). ∗p < 0.05, ∗∗∗p < 0.001 by one-way ANOVA with Dunnett's test comparing with siNC. Data represent means ± SD.