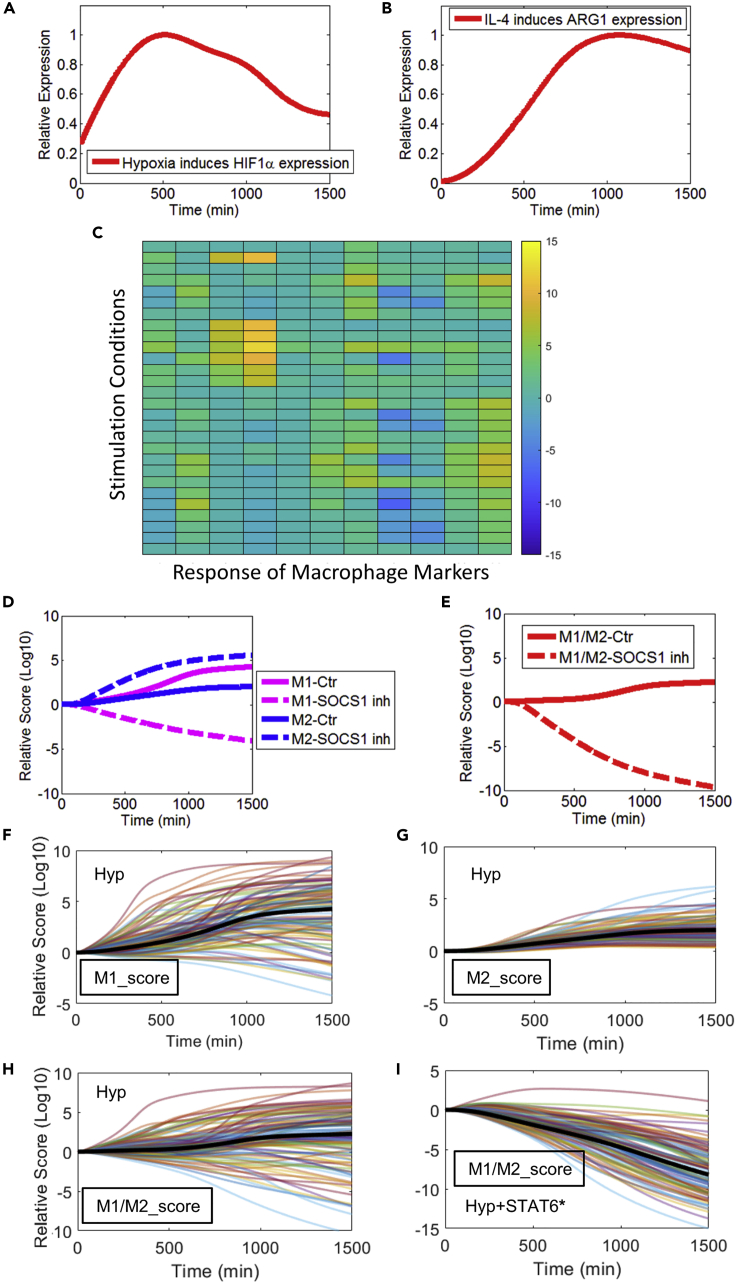
Figure 1.
Expected outcomes of simulation examples discussed in this protocol
(A and B) Steps 1–5: simulated time-course expression profiles of HIF1α (upon hypoxia) and ARG1 (upon IL-4 stimulation). Simulation results are normalized to the respective maximum values.
(C) Step 6: an example of a simulated macrophage phenotype map. Here, the values for the relative fold changes of all markers are taken at 4 h and then log2 transformed. More details of this map can be found in the MATLAB script provided.
(D and E) Steps 7–10: simulated phenotype profiles (in terms of M1, M2, and M1/M2 scores) of macrophages in hypoxia versus hypoxia plus SOCS1 inhibition.
(F–H) Steps 11–14: simulated phenotype profiles (in terms of relative M1, M2, and M1/M2 scores) of 100 model-based “virtual macrophages” in hypoxia (Hyp).
(I) Step 15: simulated phenotype profiles (in terms of the relative M1/M2 scores) of 100 model-based “virtual macrophages” in response to hypoxia and an in silico intervention targeting STAT6. (D–I) Results are normalized to their respective t=0 values and then log10 transformed.