Abstract—
Haptoglobin (Hp) is a glycoprotein that binds free hemoglobin (Hb) in plasma and plays a critical role in tissue protection and prevention of oxidative damage. Besides, it has some regulatory functions. Haptoglobin is an acute-phase protein, its concentration in plasma changes in pathology, and the test for its concentration is part of normal clinical practice. Haptoglobin is a conservative protein synthesized mainly in the liver and lungs and is the subject of research as a potential biomarker of many diseases, including various forms of malignant neoplasms. Haptoglobin has several unique biophysical characteristics. The human Нр gene is polymorphic, has three structural alleles that control the synthesis of three major phenotypes of haptoglobin: homozygous Нр1-1 and Нр2-2, and heterozygous Нр2-1, determined by a combination of allelic variants that are inherited. Numerous studies indicate that the phenotype of haptoglobin can be used to judge the individual predisposition of a person to various diseases. In addition, Hp undergoes various post-translational modifications (PTMs). These are structural transformations (removal of the signal peptide, cutting off the Pre-Hp precursor molecule into two subunits, α and β, limited proteolysis of α-chains, formation of disulfide bonds, multimerization), as well as chemical modifications of α-chains and glycosylation of the β-chain. Glycosylation of the β-chain of haptoglobin at four Asn sites is the most important variable PTM that regulates the structure and function of the glycoprotein. The study of modified oligosaccharides of the β-chain of Hp has become the main direction in the study of pathological processes, including malignant neoplasms. These characteristics indicate the possibility of the existence of Hp in the form of a multitude of proteoforms, probably performing different functions. This review is devoted to the description of the structural and functional diversity and the potential use of Hp as a biomarker of various pathologies.
Keywords: haptoglobin, biomarker
INTRODUCTION
Haptoglobin (Hp), a conservative protein that presents in all mammals, was discovered in 1938 “as a substance in blood plasma that binds hemoglobin” [1]. Accordingly, its name was formed from the combination of the words “haptein” (Greek for “to bind”) and “hemoglobin” (Hb). Нр forms an extremely strong non-covalent complex with free Hb (1 : 1 mol/mol), which protects tissues from oxidative damage [2]. Haptoglobin is multifunctional, plays an important role in various biological processes, and is currently considered as a potential biomarker of many diseases, including various forms of malignant neoplasms. Also, Hp exhibits immunoregulatory properties, participates in the inhibition of nitric oxide, stimulates tissue repair, is involved in angiogenesis, etc. The concentration of Hp in plasma changes with pathology and the test for its concentration is part of normal clinical practice.
1 HAPTOGLOBIN GENE(S)
Only human Hp is characterized by molecular heterogeneity due to genetic polymorphism. In humans, the gene Hp is located on chromosome 16 and can be present in the form of three alleles: Hp1F, Hp1S, Hp2, which control the formation of six Hp phenotypes: 1F-1F, 1S-1S, 1F-1S, 2-1F, 2-1S, 2-2, [3–5]. Due to the lack of a functional difference between Hp1F and Hp1S, which differ only in point mutations, only two alleles, Hp1 and Hp2, are often considered, which manifest themselves as three phenotypes: homozygous Hp1-1 and Hp2-2, and heterozygous Hp2-1 depending on the combination of inherited allelic variants. It is assumed that the Hp2 allele arose as a result of intragenic duplication of a 1700 bp DNA fragment of gene Hp1 after human divergence in the late evolution of primates [6].
The estimated frequency of different types of haptoglobin is as follows: Нр1-1—15–18%; Нр2-1—46%; Нр2-2 —38%, but significant geographical differences were noted. For example, in Southeast Asia, the frequency of the Hp1 allele is only about 0.1, while in other regions, it can reach about 0.8 (South America). The observed differences in the distribution of Hp phenotypes are probably the result of both genetic drift and natural selection [7]. Some methods of forensic medicine are even based on this, for example, to establish controversial paternity [8].
It should be noted that primates have two more genes derived from the gene Hp—Hpr and Hpp (Fig. 1). The Hpr gene is 90% identical to the Hp gene and is located on the same chromosome at a distance of only 2200 bp, after another 16 000 bp the Hpp gene is located. The Hpr and Hpp genes appear to have arisen as a result of the triplication of the Hp gene during primate evolution. Subsequently, the Hpp gene in humans was deleted, and only two genes remained in the current cluster of human Hp genes [9].
Fig. 1.
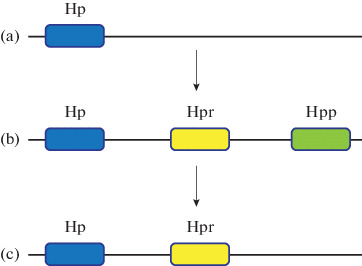
Schematic representation of the evolution of the haptoglobin gene cluster. The Hpr gene arose because of the three-fold repetition of the Hp gene after the New World monkeys moved away from other primates. Thus, new world monkeys (a), such as the spider monkey, carry only one Hp gene, whereas Old World monkeys (b), of which the chimpanzee is an example, possess the Hp and Hpr genes in addition to the so-called primate haptoglobin gene (Hpp). During human evolution (c), one locus was removed, and only 2 genes (Hp and Hpr) remained.
In humans, haptoglobin is synthesized mainly in the liver and lungs and is secreted into the blood plasma [10], but its mRNA is also detected in the kidneys, spleen, thymus, and heart [11]. The expression level of the Hpr gene is 1000 times lower than that of Hp. Hpr, like Hp, is also expressed mainly in the liver, but in addition to this, its mRNA is found in many other organs (https://www.uniprot.org/uniprot/ P00739).
Analysis of the homology of genomic sequences showed that teleosts are an utmost member of the phylogenetic series with a gene that encodes a protein homolog of mammalian haptoglobin. These results suggest that Hp evolved from a complement-associated protein (mannose-binding lectin-associated serine proteinase, MASP) with the advent of fish. The exception was the chicken and the frog, they lack this gene. However, another protein that binds Hb and is a potent antioxidant, PIT54, was identified in chicken plasma [12].
2 STRUCTURAL ASPECTS
Human haptoglobin is a tetramer of two light α‑chains (α1 or α2) and two heavy β-chains linked by disulfide bridges into αβ-dimers, and then into tetramers, and structurally resembles immunoglobulins. Characterization of the haptoglobin cDNA showed that the α-chain (α1 or α2) and the β-chain are encoded by the Нp gene in tandem form, because of which the full-length precursor of haptoglobin (pre-haptoglobin, Pre-Hp) is synthesized. According to the UniProt database, there are two isoforms of haptoglobin, or rather Pre-Hp: the canonical isoform 1 (Pre-Нр2), which consists of 406 amino acid residues (a.a.) (Mw 45 200), and isoform 2 (Pre-Нр1), which consists of 347 a.a. (Mw 38 452) and differs from isoform 1 in absence of the 38−96 amino acid region. (Fig. 2). Inside the full-length polypeptide, the α- and β‑chains are separated by one linker codon corresponding to the Arg84 α1-chain and Arg143 α2-chain [13]. Only the α-chains differ, and the β-chain (pI 6.32/Mw 27 265) is identical in three alleles (245 a.a.) [14]. Alleles Нp1S, Hp1F encode polypeptide chains α1S (slow) and α1F (fast) (pI 5.23/Mw 9192) of the same length (83 a.a.), but α1F has aspartic acid and lysine residues at positions 52 and 53, and α1S—residues of asparagine and glutamic acids in the same positions [6]. In the case of α2, additionally, the same amino acid substitutions are corresponding to positions 111 and 112 [15]. Heterogeneity of the α‑chains of haptoglobin is detected by the separation of proteins by two-dimensional electrophoresis (2DE) [15, 16]. The α1 chain has two free SH-groups of Cys residues. One is at the COOH-terminus, Cys72, and always binds to the β-chain by a disulfide bond, forming a unit (α1β-dimer). The other, at the NH2-terminus, Cys15, binds to another (α1β) unit, resulting in the formation of (α1β)2 or mature Hp1-1, which is a tetramer [15, 16]. The Hp2 allele is a product of partial gene duplication as a result of non-homologous crossing over between Hp1S, Hp1F alleles and encodes the α2-chain (pI 5.57/Mw 15 946) [15]. It contains the same a.a. as α1 and an additional copy of 12−70 a.a.— 142 a.a. in total. By binding to the β-chain, the α2‑chain forms (α2β) unit. Unlike α1, α2 has three free SH-groups of Cys residues, one of them can interact with an additional αβ unit (α1β or α2β), with the formation of large polymers (heterogeneous mixture of multimers) [13, 17, 18]. According to recent genetic analyzes, the larger Hp2 variant is considered the progenitor of all human Hp alleles. It follows from this model that exons 3 and 4 of the Hp2 allele of the progenitor were deleted due to non-allelic homologous recombination during the structural evolution of Hp2 into Hp1 [12, 19–21]. These two exons determine the multimeric structure of Hp2-2 and Hp 2-1 [22]. The Нр0-0 phenotype is characterized by the absence of Нр or its decreased level in plasma (agaptoglobinemia) and indicates that Нр, surprisingly, is not necessary for the organism’s survival. It is assumed that the decrease in the level of Hp expression is associated with possible mutations [23, 24].
Fig. 2.
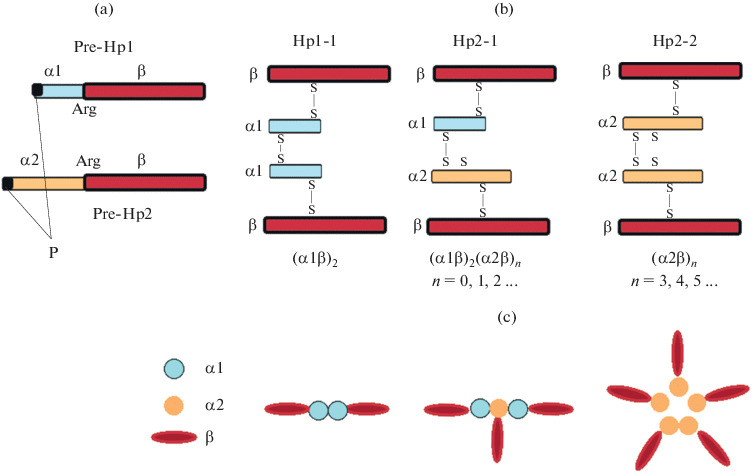
Diagram of maturation and structure of different types of Hp. (a) Pre-Hp is translated as a single-chain precursor protein; Arg, restricted proteolysis site; P is a signal peptide. (b) Assembly of α1β and α2β dimers and Hp tetramers using disulfide bonds at free Cys (S) residues. (c) Hp1-tetramer, Hp2-1-linear polymer, Hp2-2-ring polymer. Adapted from [30, 48].
Protein Нр has a domain structure. Hp variants are largely homologous to MASP, with the α-chain containing the complement control protein (CCP) domain, and the β-chain containing the catalytically inactive chymotrypsin-like serine proteinase (SP) domain [25]. This is an amazing example of homologous proteins with different biological functions. Active sites typical of serine proteases (His57 and Ser195) in Hp are replaced by lysine and alanine, respectively, which excludes the possibility of Hp protease activity [26].
Many secreted proteins are synthesized as pro-forms and become biologically active after proteolytic cleavage (processing) in the Golgi apparatus. Hp is unusual in that it is cut in the endoplasmic reticulum before entering the Golgi apparatus [13, 27, 28]. Haptoglobin is transcribed and translated as a single polypeptide, a precursor of either Pre-Hp1 or Pre-Hp2, depending on the genotype/phenotype. During processing, the N-terminal signal peptide is first removed from it, and then it undergoes complex post-translational modifications in the endoplasmic reticulum. At the site of restricted proteolysis (Arg143 in the case of Hp2 and Arg84 in the case of Hp1), the polypeptide splits into two subunits: the N-terminal light α-chain and the C-terminal heavy β-chain. The C-terminal Arg of the α-chain is removed by carboxypeptidase N. Both chains are covalently linked by a disulfide bond, forming the basic α1β or α2β unit (dimers) [19, 20] (Fig. 2). Interestingly, a variant is possible here when the canonical form encoded by the Hp2-2 allele does not undergo proteolytic processing, remains full-length, and functions as a zonulin protein. Proteomic analysis showed that the zonulin protein is identical to Pre-Hp2 [29]. Zonulin is produced in the intestinal mucosa and in the liver, reversibly regulates intestinal permeability, modulating tight junctions between the cells of the digestive tract wall, and controls the balance between tolerance and immunity to foreign antigens. Several diseases are associated with an increased level of zonulin: autoimmune, inflammatory, and neoplastic [30] (Fig. 2). Being in the intact full-length form of the precursor protein, zonulin acts to modulate the permeability of the intestinal barrier, while the main function of mature Hp is to form a stable complex with Hb and prevent hemoglobin-induced oxidative tissue damage. This is a striking example of the dependence of the function of a protein on its structure. The importance of the Hp conformation is emphasized by the fact that antibodies to zonulin identify the α1-chain of Hp only under denaturing conditions [30].
Thus, proteins with different structures, molecular weights, and molecular heterogeneity correspond to the three major Hp phenotypes. According to the results of native electrophoresis, it follows that Нр1-1 (Mw 86 000) is a tetramer (α1β)2; Hp2-1 (Mw 86 000–300 000) and Hp2-2 (Mw 170 000–900 000) are heterogeneous (multimeric) polymer mixtures (Fig. 3). Full forms of haptoglobin of different types consist of (αβ) units with different numbers of thiol groups in α chains (cysteine residues): in α1β there are two cysteine residues and in α2β—three. As a result, depending on the phenotype, there are different structures of haptoglobin: Нр1-1—(α1β)2; Нр2‑1—(α1β)2 (α2β)n, where n = 0, 1, 2, 3…; Нр2‑2—(α2β)n, where n = 3, 4, 5… . Forms with 10 repeating units (αβ)10 in Нр2-1 and 20 repeating units (αβ)20 in Hp2-2; Нр1-1 has one form corresponding to the αβ‑dimer [18, 31–33] (Fig. 3). Besides, it follows from these studies that haptoglobin with the Нр2-1 phenotype is a linear polymer in which α1β always forms terminal units, and with the Нр2-2 phenotype, it is a ring polymer, a cyclic structure consisting of many (α2β) units. These structures of haptoglobin are confirmed using electron microscopy. The functional role of such complex structural HP variants is still unclear [13]. An individual belonging to the Hp polymer phenotype is associated with different levels of the antioxidant function of Hp and the risk of many common diseases [34–36].
Fig. 3.
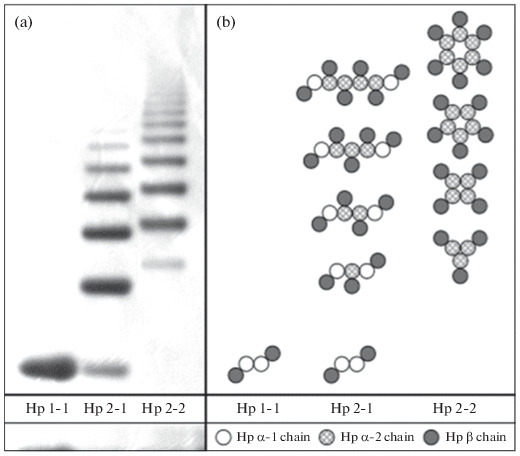
Assessment of the phenotype of haptoglobin using native PAGE. (a) Specific profiles obtained by electrophoresis in gradient (3-8%) native PAGE of haptoglobin preparations of various phenotypes. (b) Composition of polymers of three haptoglobin phenotypes: Нр1-1—homodimers; Нр2-1—linear heterodimers; Нр2-2—cyclic heterodimers. Adapted from [31].
3 BLOOD PLASMA PROTEINS AND THEIR VARIABILITY
The functioning of haptoglobin cannot be considered in isolation from the entire plasma, of which it is one of the important components. Plasma proteome is a dynamic, complex mixture of proteins, an important substance for clinical analysis. Proteomic profiles of plasma proteins can illustrate differences associated with metabolic changes or pathological processes since many proteins circulating in the plasma are products of cascades of enzymatic reactions of all tissues and organs and can reflect the physiological state of the body. Systemic proteomic profiling of most of the circulating plasma proteins (plasma proteome) makes it possible to detect new biomarkers to predict and diagnose diseases [37]. For example, the use of proteomic approaches to compare proteomes in patients with glioblastoma and healthy donors have shown that some serum proteins associated with different vital physiological pathways, including haptoglobin, are represented in different ways [38].
Analysis of plasma proteins is difficult due to the complexity of the plasma proteome, its dynamism, chemical similarity of many proteins, and a huge range of concentrations of various proteins [39]. Acute-phase proteins are common indicators and biomarkers for prognostic and diagnostic assessment of the state of the body. The acute-phase response of homeostasis disturbance produces a cascade of changes in the regulation of transcription, which leads to an increase in the synthesis of some proteins and a decrease in the synthesis of others [40]. Prominent examples of an increased protein response are C-reactive protein, serum amyloid A, and haptoglobin. These proteins are produced in the liver as a result of induction by cytokines during inflammatory processes and are secreted into the plasma [41]. It should also be noted the importance of assessing the variability of the proteomic profile of a potential marker protein in healthy individuals of the control group before carrying out comparative proteomic analyzes to distinguish changes characteristic of any disease from the natural plasticity of the proteome. The variability in the level of some major plasma polypeptides can be comparable or even greater than the difference in levels found in pathology [42]. In the case of haptoglobin, its level is normally in the range of 450 mg/L to 1650 mg/L. A level below 450 mg/L may indicate an increased rate of red blood cell death. The increased breakdown of red blood cells leads to an increase in the supply of hemoglobin into the blood and, accordingly, to a decrease in the level of haptoglobin. Therefore, the determination of the content of haptoglobin is used primarily to identify and assess the degree of hemolytic anemia. For individual Hp chains, the concentration ranges in healthy individuals are as follows: for the β-chain, 6–40 μM; for the α1-chain, 0–40 μM; for the α2‑chain, 0–40 μM [43].
The complexity of the canonical structure of haptoglobin (homodimers and heterodimers, structured into tetramers); the formation of heterogeneous linear and cyclic polymer structures (polymorphic variants of the α1-chain (α1F, α1S) [16], proteolytic processing and PTM of α1- and α2-chains, the presence of four sites of active glycosylation of the β-chain) indicate that haptoglobin can exist in the form of a variety of proteoforms and to be involved in the performance of various functions [33]. Also, individuals are often burdened with several diseases, each of which can contribute to the change in the level and nature of haptoglobin’s PTMs, so it is always necessary to examine a large amount of data to obtain statistically reliable results. From the above, it follows that Hp is not a very suitable protein as a possible biomarker based on the assessment of changes in the total level of this protein only. However, with the help of modern proteomic technologies, among thousands of protein forms (proteoforms), those that undergo significant modifications in pathology are found, allelic variants and variable PTMs are identified by changes in the fundamental physicochemical characteristics of the protein: molecular weight (Mw) and isoelectric point (pI). An important point here is phenotyping, which is the possibility of a clear definition of the type of Hp of an individual and the constancy of this characteristic throughout life [33, 35, 44]. Due to this, Hp can be an extremely useful nonspecific polypeptide, a potential marker of many, including malignant neoplasms. Knowledge of the many plasma proteoforms of Hp can be important for the strategy, diagnosis, treatment, and prevention of various diseases, that is, it has great therapeutic potential.
There is evidence that in some cases the unprocessed Prе-Нр form (Fig. 2) may be a more useful candidate for a biomarker than the mature Нр. For example, in the serum of patients with hepatoma, the Prе-Hp level is much higher than in healthy donors [45].
4 BASIC FUNCTIONS AND ROLE OF THE Hp PHENOTYPE
Numerous studies indicate that haptoglobin circulates in the blood plasma as a polymer, whose stoichiometry, size, and biophysical properties depend on three main, hereditarily determined phenotypes (homozygous Hp1-1, Hp2-2, and heterozygous Hp2‑1), and are fundamentally different. Moreover, structural differences between phenotypic forms have important functional consequences [46].
The most important function of haptoglobin is the ability to bind free hemoglobin, to be an antioxidant. The Hp bond with free Hb (complex) has the highest protein-protein affinity with Kd = 10–14 M [47]. This underscores the importance of the biological process under consideration for protecting the body from oxidative damage. It was established by the method of selective proteolysis that only heavy β-chains of Hp are involved in binding [48]. Significant negative effects occur within tissues when hemoglobin remains in circulation. Free hemoglobin is highly toxic due to the oxidative nature of heme, which contains iron involved in the production of reactive oxygen molecules that cause cell damage [14, 49]. Moreover, it was found that the ability to reduce the degree of damage caused by free radicals depends on the Hp phenotype [50]. Numerous studies indicate that an individual’s belonging to the Hp polymeric phenotype (Hp2-1 and, especially, Hp2-2) is associated with the risk of many common diseases: cardiovascular, autoimmune, infectious, atherosclerosis, stroke, diabetes, tuberculosis, epilepsy, diabetic nephropathy, diabetic retinopathy, malignant neoplasms, weakening of cognitive functions [33–36, 51–53]. However, in the case of prostate cancer, the role of the phenotype has not been found [54]. An important site of Hp synthesis, apart from the liver, is the lungs. Haptoglobin protects the lungs from inflammatory agents and prevents lung damage. Its deficiency leads to the destruction of lung tissue caused by the natural bacteriostatic function of neutrophils, which leads to emphysema and chronic obstructive pulmonary disease (COPD). In [10], the Нр2-2 phenotype is considered a new biomarker for COPD identification. The determination of the patient’s haptoglobin phenotype may be an important factor in the prognosis of the disease and the course of treatment [35]. Note that in routine laboratory practice for phenotyping Hp, a method based on a PCR reaction taking into account the structural differences of Hp1 and Hp2 alleles is successfully used [24]. Another new method of Hp phenotyping using fluorescent probes of gold nanoclusters (AuNC) is described in [55].
A pathological situation affecting the level of Hp in the blood is an acute phase condition, which includes infection, inflammation, tumor, burn, frostbite, chemical damage, autoimmune disease, hemolysis, etc. Here, the main function of Hp is to bind free hemoglobin released from destroyed red blood cells. Hemoglobin is contained in red blood cells, delivers oxygen to tissues, and participates in the transport of carbon dioxide. The lifetime of an erythrocyte is 120 days. The release of hemoglobin into plasma is a physiological phenomenon associated with intravascular hemolysis due to the destruction of old erythrocytes at a rate of 2 × 106 cells/s [56, 57]. Free Hb binds to Hp, forming a Hp/Hb complex. In the liver, the complex is delivered to the reticuloendothelial system by endocytosis mediated by the CD163 receptor and is degraded. In this way, the loss of free hemoglobin is reduced, iron is recycled (it is returned to the resulting erythrocytes) and the antioxidant function is performed to protect the kidneys from damage by toxic radicals.
In in vitro experiments using mass spectrometry and plasmon resonance, a significant difference in the binding capacity of the two types of haptoglobin, Hp1-1 and Hp2-2, with hemoglobin was not found [58]. When determining the effectiveness of individual phenotype-specific Hp drugs in vivo experiments on guinea pigs with direct infusion, both types of haptoglobin show the same therapeutic efficacy under hemolytic conditions, with hypertension, and effectively prevent the oxidation of LDL (low-density lipoproteins) by hemoglobin. That is, both therapeutic drugs (Нр1-1 and Нр2-2) bind free Hb equally well with high affinity preventing iron loss and protecting tissues from oxidative effects, even though Нр2-2 has a heterogeneous multimeric structure compared to dimers Нр1-1. In a review [56], the authors outline possible mechanisms that provide the difference in the functions of Hp of different types. These studies are important for elucidating the efficacy of Hp preparations of a particular phenotype, and the results presented in the article do not support the concept that phenotype-specific Hp therapists have different efficacy in mitigating hemoglobin toxicity. The authors of the review [56] believe that all three Hp phenotypes have the same binding capacity with Hb, but Hp1-1 has a greater ability to protect against oxidative damage. It should also be noted that there is a publication indicating that the concentration of Hp1-1 in plasma is higher than the concentration of Hp2-1 and Hp2-2 [59]. However, it was shown in [60] that the oxidation of LDL, which plays an important role in atherosclerosis, is practically inhibited by Hp1-1 and only partially by Hp2-2. Further research on this issue appears to be needed. Note that in Japan, Hp preparations under acute hemolytic conditions have been clinically used since 1985 [61].
It was shown that Hp is an extremely powerful antioxidant that directly protects low-density lipoproteins from Cu2+-induced oxidation [14]. The potential of Нр is noticeably higher than probucol (one of the most powerful antioxidants). The level of antioxidant activity is distributed in the following order: Hp 1-1 > Hp 2-1 > Hp 2-2 > probucol > vitamin E.
In an in vitro system, on fibroblasts of rabbit arteries, it was shown that Hp is produced in arterial tissue, where it is involved in cell migration and vascular restructuring [62]. Usually, about 1% of erythrocytes are destroyed and removed from circulation per day. However, an increase in this amount to at least 2% leads to the complete disappearance of haptoglobin from the blood [26, 63]. Decreased Hp level is a sign of intravascular hemolysis induced by malaria, poisons, toxins since in the presence of a large amount of free hemoglobin, haptoglobin is depleted [11, 64–66]. However, it is noted that a decrease in the Hp level can also occur in the absence of hemolysis, for example, due to liver cirrhosis, disseminated ovarian carcinomatosis, pulmonary sarcoidosis, or increased estrogen levels [11]. With inflammation, tumor growth, and chemical damage, the level of haptoglobin in the blood rises [41]. Hp has also immunoregulatory properties. It plays a modulating role in the balance of Th1/Th2 helper cells, which are characterized by different patterns of cytokine expression after activation and are responsible for the immune response [67].
To date, fundamentally important results on the role of haptoglobin have been obtained in numerous studies of diabetic patients. First, the incidence of diabetes in individuals with type Hp2-2 haptoglobin is significantly higher than with type Hp1-1 [34, 68]. This disease is characterized by cardiovascular, nervous, and infectious complications. They are largely associated with oxidative stress and antioxidant defenses. Persons with diabetes and with the Hp1-1 phenotype are resistant to the development of diabetic retinopathy, diabetic nephropathy, and cardiovascular diseases [56]. The authors of the publication [56] believe that the type of Hp should be considered in diabetic patients as an additional risk factor for cardiovascular diseases. In diabetic patients with Нр2-2, this risk is five times higher, and with Нр2-1, it is three times higher than in patients with Нр1-1 [49]. Three possible mechanisms of different Hp behavior depending on the phenotype are discussed. First, molecular differences in the structure and size of haptoglobin of different types (steric factor) can prevent or exclude the entry of oligomeric molecules of haptoglobin Нр2-1 and Нр2-2 into the intravascular space, where Нр binds and neutralizes free Hb. As a result, in individuals with the Нр2-1 and Нр2-2 phenotype, free hemoglobin remains circulating for a significant time, contributing to the transition of oxidative stress to vascular stress [49]. Secondly, glycation of hemoglobin is an additional factor that reduces the effectiveness of Нр2-2 as an antioxidant. Diabetes is characterized by the fucosylation of many proteins, including hemoglobin. It was shown that the oxidizing activity of glycated Hb is not effectively blocked by haptoglobin, and there is a difference in the rate of removal of complexes of glycated Hb with haptoglobin by hepatocytes as compared to the control [68]. It was found that the risk of coronary heart disease increases in patients with Hp2-2 when the degree of glycation of hemoglobin is >6.5% [69]. That is, a mechanism of significant dysfunction of Нр2-2 with glycated hemoglobin is possible [70]. Third, the presence of the Нp2 allele in diabetes may be associated with an increase in the amount of active iron [71]. The difference in the binding affinity of Hp1-1 and Hp2-2 with hemoglobin has not been detected, but it was found that Hp1-1 and Hp2-2 differ in their ability to prevent the transfer of iron from hemoglobin to the chelated active form. A noticeable increase in the amount of active iron was shown in the case of the Нр2-2/Hb complexes. In the plasma of genetically modified mice producing Hp2-2, a diabetic-dependent difference in the amount of active iron was also identified. These experimental results indicate that active iron is involved in increasing the susceptibility of individuals with the Нp2 allele to vascular diseases in diabetic patients, and it follows that, in the context of diabetes, the role of haptoglobin can be both protective and pathogenic.
Iron is one of the most important elements of bacterial growth. The combination of hemorrhagic injury and infection can be fatal due to the presence of blood in the damaged tissue, which provides the necessary iron for the development of microorganisms [72]. However, once bound with haptoglobin, hemoglobin and iron are no longer available to bacteria. Indeed, it has been shown that the negative consequences of intraperitoneal administration of E. coli and hemoglobin to rats can be prevented by the administration of haptoglobin [72]. In the mucous layer and alveolar fluid of the lungs, haptoglobin is the main component of antimicrobial activity and plays an important role in protecting against infection [44, 73].
Individuals with type Hp2-2 show high immunological reactivity, including high production of antibodies after vaccination. It was found that Hp inhibits the synthesis of prostaglandins and thus exhibits anti-inflammatory properties [67, 74].
Haptoglobin is involved in the inhibition of nitric oxide. Nitric oxide is involved in maintaining vascular tone and is largely depleted by both, hemoglobin and Hb/Hp complex, which is undesirable in the case of cardiovascular diseases. The function of inhibition of nitric oxide is better performed in the case of Нр1-1 since the Нр1-1/Hb complex is more efficiently removed from the circulation as compared to the Нр2‑2/Нb complex [75].
In several works, the angiogenic activity of mature Hp and Prе-Hp isoforms has been demonstrated. In an in vitro system, it was shown that Prе-Нр enhances the expression of growth factor and receptor (VEGF/VEGFR2), as well as processes and branches in endothelial cells that line the inner surface of blood and lymphatic vessels, that is, stimulates angiogenesis. Moreover, mutant Prе-Нр that cannot turn into Нр due to a point mutation (replacement of Arg143 (CGG) with glutamine (CAG) appears to be more effective [45]. In patients with systemic vasculitis, Hp stimulates tissue repair and compensates for ischemia by promoting the growth of additional vessels. Moreover, Нр2-2 was found to be more angiogenic than other types of Hp [34, 56, 76]. In an in vitro system, on fibroblasts of rabbit arteries, it was shown that Hp is expressed in arterial tissue and is involved in cell migration and vascular restructuring [62]. The question of whether Hp belongs to the group of chaperone-like proteins, the so-called extracellular molecular chaperones, capable of stabilizing the conformation of the protein and protecting it from aggregation under stress conditions, is also being investigated. For all types of Нр, prevention of acidic (Н2О2) and thermally induced aggregation of catalase and γ-crystallin (eye lens protein) and only partial refolding of these proteins has been shown [77]. In experiments using fatty acids (oleic, stearic, palmitic), it was shown that the Hp2-2 form prevents the formation of β2-microglobulin fibrils, which cause inflammation during prolonged dialysis. The formation of amyloid fibrils in such experiments was completely inhibited [2].
It was found that patients with bronchial asthma, which having the Нр2-2 phenotype, are characterized by more pronounced immunological reactivity in comparison with persons with other phenotypes. That is, it can be assumed that the Hp2-2 phenotype is an associated biological marker of bronchial asthma [78]. There is literary evidence that people with the Hp2-2 phenotype are more prone to systemic sclerosis than those with the Hp1-1 phenotype. Moreover, proteomic analysis indicates an increased level of one of the α2-proteoforms in patients with systemic sclerosis [79].
In the light of recent epidemic events, studies have been conducted to identify the causes of the difference in spread and mortality from COVID-19 in different geographic regions. In particular, this could be associated with a strong geographic variability of plasma protective proteins, including Hp [80, 81]. The different phenotypes of haptoglobin did not show a significant correlation with the prevalence of COVID-19 [82]. However, for another protein, ACE1 (angiotensin-converting enzyme 1), a strong correlation was found in its polymorphism, which can be considered as a factor influencing the spread of COVID-19 and the outcome of infection [82].
The haptoglobin-related protein Hpr is also secreted into plasma, where it is bound to apolipoprotein L-I (apoL-I), which contains high-density lipoprotein (HDL). This complex is called trypanosomal lytic factor-1 (TLF-1) since it provides innate immune protection in humans against many species of African trypanosomes [83]. Also, Hpr, like Hp, can bind hemoglobin with high affinity and assist in the clearance of free hemoglobin to ensure the recirculation of heme iron in the liver. Moreover, in contrast to the Hp/Hb complex, the Hpr/Hb complex does not have an increased affinity for the CD163 receptor; therefore, the mechanisms of Hb utilization, in this case, should be different [83].
Despite the weakened functions, the Нр2 allele dominates in the human population (up to 80% in some ethnic groups) [18]. There are also reports of the predisposition of persons belonging to the Hp1-1 group to many diseases, in particular to leukemia. So, according to some data, the likelihood of acute lymphocytic leukemia in people with the Hp1-1 group was significantly higher than in people belonging to the Hp2-2 group. The authors believe that the Hp1-1 phenotype can be considered as a sign of an increased risk of chronic lymphocytic leukemia [84]. However, the authors of another study did not find an increase in the frequency of Hp1-1 representation in patients with chronic lymphocytic leukemia and conclude that in these patients the location of the Hp fractions on the electrophoregram may change and lead to errors when considering the Hp groups [85].
5 HAPTOGLOBIN AND ITS PROTEOFORMS AS POTENTIAL BIOMARKERS OF DISEASES
A wide variety of Нр proteoforms arises due to processing and PTMs. Especially many additional protein variants are formed during glycosylation of the β‑chain of haptoglobin at four Asn sites (Asn184, Asn207, Asn211, Asn241) [32, 86]. Numerous studies have shown that changes in the modified oligosaccharides of the β-chain of Hp in serum/plasma occur in various pathologies, including malignant neoplasms. This makes Hp (along with other glycan markers) a promising, non-invasive biomarker for diagnosing, predicting, and monitoring patient treatment.
Many studies focus on the identification of PTMs and changes in the level of the Hp α2-chain in pathology [15, 39, 87]. These characteristics of Hp indicate the possibility of this protein existing in the form of various proteoforms. Their identification is important for understanding the physiological and pathological role of Hp.
Proteomic technologies are widely used to study PTMs in blood plasma in the case of various diseases. The main research methods for the identification and visualization of proteins are one-dimensional denaturing sodium dodecyl sulfate electrophoresis (SDS-PAGE) and two-dimensional electrophoresis (2DE) in combination with Western blotting or mass spectrometry (MS). Such experiments aim to obtain a protein profile (pattern) representing a combination of various proteoforms (Fig. 4). Comparison of 2DE-immunoblots of protein in normal and pathological conditions can give a picture of changes in such parameters as the number of proteoforms (protein spots), their intensity, size, and position. And with the help of MS, numerous site-specific modifications are identified. As for haptoglobin, according to the PhosphoSitePlus information resource, there are 5 serine and tyrosine phosphorylation sites, 5 lysine acetylation sites, and one lysine ubiquitination site of the human Hp (phosphosite.org). The characteristic protein profile of the α2-chain of Hp in 2DE-gels upon Coomassie staining is three spots with practically identical coordinates by weight, but different in pI (Fig. 4). The central major spot most likely corresponds to the unmodified form, which has the amino acid sequence shown in the Uniprot database as P00738; one spot migrates to a more acidic pH range (deamidation of Asn residues at position 5 to aspartic acid); the other spot is displaced to the alkaline side due to the presence of an additional C-terminal residue Arg (basic amino acid), a linker between the α‑ and β-chains. This information on PTMs of Hp was obtained using the MS and published in [15]. In [39], a change in the profile of these proteoforms in plasma during pregnancy with fetal growth restriction (FGR) was found. These three forms of α2-Hp showed different patterns in FGR patients and healthy individuals, and the presence of a specific α2-Hp variant unique to FGR patients was found. That is, haptoglobin is a potential marker of this pathology, the prevalence of which is 3% and poses a significant threat to the health of patients. Also, a change in the protein profile and a significant decrease in the level of β- and α-chains of Hp in the plasma of patients with malaria compared with healthy donors have been shown [89]. According to available data [90], phosphorylated and glycosylated proteoforms of the haptoglobin α2-chain are detected in the plasma of patients with Non-Small Cell Lung Cancer.
Fig. 4.
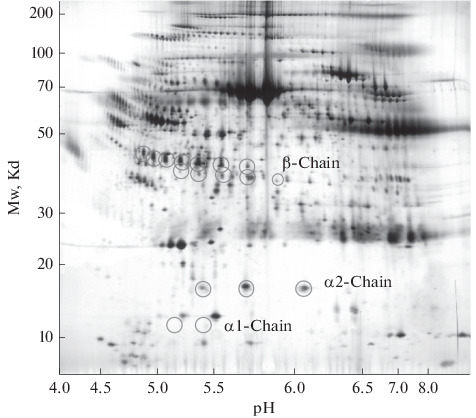
The 2D electrophoretic map of human plasma proteins. The circles indicate the position of the haptoglobin proteoforms. Adapted from [88].
Interesting results were obtained, when analyzing the sera of patients with squamous cell lung cancer [91]. 2DE showed an increased level of β-chain proteoforms. Moreover, in MS analysis, two peptides of these proteoforms were especially pronounced. These data were also supported immunologically. The authors of the publication [91] believe that the detection of one of these peptides (peptide HP216) is a promising biomarker for the early detection of squamous cell carcinoma, especially in combination with the determination of soluble fragments of cytokeratin 19 (CYFRA 21-1), used for diagnosis, prognosis, and control treatment of lung cancer, as well as some other malignant neoplasms [91, 92].
In patients with ovarian cancer, a significant increase in the level of Hp in plasma was found in comparison with the control, and haptoglobin was recognized as an independent prognostic marker of this disease [93]. Moreover, the increased amount of fucosylated forms of the α-chain of haptoglobin in ascitic fluids and tumor tissues of patients with ovarian cancer confirms their potential as biomarkers of disease progression [94, 95].
In the case of breast cancer, it has been shown that the level of Hp is increased not only in the blood but also in tumor tissues. Moreover, it was unexpectedly found that there is a functional relationship between Hp and glycolytic enzymes, such as glucose-6-phosphate isomerase and hexokinase [96]. Another disease recently found to have elevated plasma Hp levels is acute myocardial infarction in young adults [97].
It should also be noted that it is possible to use not only plasma but also saliva for the detection of tumor markers [98], including haptoglobin [99]. For example, comparative proteomic analysis revealed a significant excess (2.5 times) of the level of haptoglobin in saliva in patients with kidney cancer compared with healthy donors [99]. The authors of this study conclude that their experimental results indicate the possibility of using haptoglobin along with the S100A9 protein as potential non-invasive biomarkers for the diagnosis of kidney cancer.
Interesting results were obtained when studying the possible relationship between the Hp level and liver metastases in colorectal cancer [100]. In this work, the authors showed that knockdown of the Hp gene by specific RNAs significantly inhibited tumor growth and invasive properties. The authors believe that an increased level of Hp can be considered in this case as a predictive marker of liver metastases [100].
In the study of plasma of patients with glioblastoma, an increase in the level and change in the protein profile of the α2-chain of haptoglobin was found. Three α2-proteoforms were identified in the control and six α2-proteoforms in patients. That is, the identification of specific forms of α2-Hp in blood plasma can be a biomarker of glioblastoma [38, 101] (Fig. 5). In our studies, this conclusion is also confirmed. Thus, according to our unpublished data, the level of the α2-chain of haptoglobin in the blood plasma of patients with glioblastoma is significantly increased in comparison with the plasma of healthy donors. Moreover, comparative analysis of 2DE-profiles revealed variability in the sets of proteoforms (patterns) of the α2-chain of haptoglobin in patients with glioblastoma. Taking into account the results obtained in [102], it can be assumed that the proteoforms of the Hp α2‑chain with a shift of the pI to the acidic region may not only be deamidated, but also acetylated, and could be potential markers of glioblastoma. Glycosylation is one of the most common and important PTMs that create numerous protein variants due to oligosaccharide heterogeneity, which affects the physical (solubility, stability), structural (folding), and functional properties of proteins [32, 86]. Changes in glycosylation, minor and significant, are associated with many diseases. Moreover, most of the plasma proteins are glycosylated. Numerous studies show that a comparative analysis of N-glycan profiles using mass spectrometry can allow early diagnosis of different types of malignant neoplasms since certain oligosaccharide structures dominate in different types of this pathology and can be used as biomarkers [103, 104]. Glycan analysis is a study of the oligosaccharide portion of glycoproteins and may include analysis of the whole glycoprotein; separation and detection of glycoforms; analysis of glycopeptides obtained after enzymatic treatment of the glycoprotein; analysis of cleaved glycans obtained after chemical or enzymatic processing of a glycoprotein. The most characteristic violation of glycosylation in comparison with the norm is the degree of branching of N-glycan chains: bi-, tri-, and tetra-antenna structures. They also differ in the number of glycosylation sites and the degree of fucosylation and sialylation [32]. The tri- and tetra-antenna variants are called Lewis Y-tipe structures. The concept of protein and site-specific microheterogeneity is introduced, which controls the structure, function, and interaction of proteins [86, 105]. Most types of malignant neoplasms are characterized by an increase in the level of haptoglobin [106]. In work [107], using 2DE and MS, it was shown a significant increase in the level of modified forms of Pre-Нр in the serum of patients with breast cancer, compared with healthy donors.
Fig. 5.
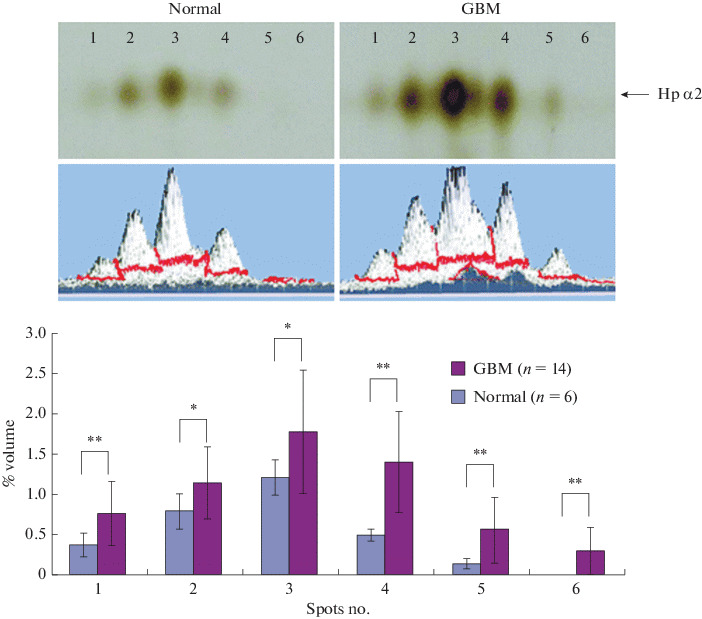
Proteomic analysis of serum from healthy subjects and patients with glioblastoma (GBM). (Top) Serum proteins from healthy donors and GBM patients were separated by 2DE and visualized by silver staining. (1−6) Spots of the α2-Hp chain with the same mass and different pI. (Bottom) Statistical analysis of spot size based on 6 (normal) and 14 (GBM) gels. Taken from [101] with permission from the American Chemical Society.
It is known that the normal liver fucosyltransferase activity is low, and most of the haptoglobin in healthy individuals is not fucosylated [108]. Studies have shown that changes in the structure of serum (plasma) blood glycans, including haptoglobin, that is, aberrant glycosylation, are often found in connection with cancer, for example, ovarian cancer [94], rectal cancer [106], cancer liver [32, 86, 104], lung cancer [109], pancreatic cancer [108], and stomach cancer [110, 111]. In the plasma of patients with pancreatic cancer, studies have shown an increase in di-, tri-, and tetra-branched glycans, and the number of triantennal glycans containing X-type Lewis fucose markedly increased in the Asn211 site of the Hp β-chain [108]. High levels of fucosylated Hp have also been found in the plasma of patients with various carcinomas [112]. An analysis of glycopeptides and cleaved permethylated N-glycans was performed in [86]. Site-specific glycoforms of haptoglobin were found with an increase in β-chain fucosylation up to six fucoses in liver diseases (cirrhosis and hepatocarcinoma). The authors believe that the observed changes are large enough and specific enough to be a diagnostic test for non-invasive monitoring of the course of the disease. In work [111], the analysis of aberrant glycosylation of Hp in blood serum in patients with gastric cancer was successfully carried out. The study [104] presents a highly sensitive and specific model for the diagnosis of hepatocarcinoma, which consists of a systemic analysis of serum Hp concentration and a comparative quantitative analysis of sialylated and fucosylated forms of haptoglobin at different stages of hepatitis and hepatocarcinoma. Using lectin-affinity purification (using glycosyl-specific lectins) and 2DE, 18 variants of S-Hp and F-Hp glycoforms were detected. Using MS, unique patterns of Hp glycoforms were found in HCC patients, consisting of hypersialated fucosylated and hyposialated fucosylated proteoforms. In special cases, the binding of fucose to the core N‑acetylglucosamine residue and the formation of tetraantennal β-chains have been shown. An increase in the level of fucosylated haptoglobin was found in a comparative analysis of the plasma of patients with psoriasis and healthy individuals. Aberrant glycoforms can reflect changes in Hp function in the skin and be used as a marker of the disease [113]. A significantly increased level of fucosylated Hp (F-Hp) in the serum of patients with pancreatic cancer was also shown, and even monoclonal antibodies highly specific to the form of F-Hp present in the patient were obtained [114, 115].
A detailed characterization of Hp glycosylation in different types of malignant neoplasms with a description of the methodological techniques used is given in the review [116]. Important results were obtained in the study of glycosylation of haptoglobin in connection with the affinity of its binding to hemoglobin. Using haptoglobin Hp1-1 as an example, it was shown that fucosylation significantly stabilizes the Hp/Hb complexes, and the (branched) antenna structures reduce the binding affinity of such complexes [105]. A large contribution is made by the data presented in [117]. Using native PAGE, gel filtration, ion-exchange chromatography, and advanced MS technologies, the authors have identified hundreds of haptoglobin proteoforms/glycoforms and their combinations. It is shown in detail how they coexist in blood serum, how they differ in individuals with different genotypes (Нр2-1 and Нр2-2) and different oligomeric states of the protein. The authors observed a correlation between the level of glycosylation and the size of the Hp oligomer. It is noteworthy that the level of fucosylation and branching of glycans decreases with an increase in the size of both Нр2-1 and Нр2-2 oligomers. According to the authors of the publication [117], oligomers with a lower mass have a higher affinity for Hb compared to oligomers with a larger mass. It is concluded that both significant fucosylation and branching of glycan structures enhance the Hp-Hb bond of the complex. Interestingly, it was shown in [105] that Hp1-1 with a higher degree of branching, on the contrary, showed a lower binding affinity with Hb. Also, the effect on the bond in the proteolytic processing complex of the Hp α-chain was found: the larger the oligomer, the higher the level of HpαR isoforms (R is the C-terminal residue of arginine, the linker between the α- and β-chains). Removal of C-terminal arginine is considered as the process of Hp maturation [15]. It was found that, in healthy individuals, the glycosylation sites Asn184, Asn207, Asn241 have preferentially sialylated biantennal N-glycans, while the Asn211 site is occupied by sialylated triantennal N‑glycans. Thus, the fine-tuning of the Hp/Hb complex formation is influenced by the cumulative effect of many factors, including the haptoglobin genotype, heterogeneous glycosylation patterns, and post-translational proteolytic processing of the haptoglobin α‑chain. It should be noted that both groups of authors, [105] and [117], used proprietary hemoglobin and haptoglobin preparations. It is obvious that the study of the glycosylation of proteins, including haptoglobin, has become one of the main directions in the research of malignant neoplasms. Glycoproteomics is a technological breakthrough that has begun to show promising results in the identification of biomarkers for early diagnosis and therapy. According to a review [118], there is growing recognition of the enormous potential of glycans for personalized medicine. However, it should be borne in mind that malignant transformation is a heterogeneous disease, and a panel of markers can provide more accurate information than a single marker [109].
CONCLUSIONS
Thus, the level of Hp in blood plasma can be used not only as a biochemical indicator for diagnosing the development of hemolytic and inflammatory processes in the body and the functional state of the liver. Also, by the phenotype of Hp, one can judge the individual predisposition of a person to various diseases. Therefore, along with the blood group and Rh factor, the phenotype of Hp is an important individual trait. When developing, introducing methods of personalized medicine and the appearance of health passports in the future, the Hp phenotype will certainly need to be taken into account. Moreover, there are grounds for the possibility of using Pre-Hp2 (zonulin), as well as proteoforms of both α-chains and β-chains of haptoglobin, as biomarkers of many, including oncological, pathologies.
FUNDING
The study was carried out within the framework of the State Assignment of the National Research Center Kurchatov Institute-PNPI for 2021−2023.
COMPLIANCE WITH ETHICAL STANDARDS
All authors declare no potential conflicts of interest warranting disclosure in this article. This article does not contain any research involving humans or the use of animals as objects.
ADDITIONAL INFORMATION
The article was translated by the author (S.N. Naryzhny).
Footnotes
Abbreviations used: a.a.—amino acid residues; CCP—the complement control protein; COPD—chronic obstructive pulmonary disease; 2DE—two-dimensional electrophoresis; Hp— haptoglobin; Hb—hemoglobin; LDL—low-density lipoproteins; MASP—mannose-binding lectin-associated serine proteinase; MS—mass spectrometry; PTM—post-translational modification; Pre-Hp—pre-haptoglobin; TLF-1—trypanosomal lytic factor-1.
REFERENCES
- 1.Polonovski M.J.M. C. R. Seances Soc. Biol. Fil. 1938;129:457–460. [PubMed] [Google Scholar]
- 2.Sultan A., Raman B., Rao C.M., Tangirala R. J. Biol. Chem. 2013;288:32326–32342. doi: 10.1074/jbc.M113.498337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Straten A., Herzog A., Cabezon T., Bollen A. FEBS Lett. 1984;168:103–107. doi: 10.1016/0014-5793(84)80215-X. [DOI] [PubMed] [Google Scholar]
- 4.Maeda N., Smithies O. Annu. Rev. Genet. 1986;20:81–108. doi: 10.1146/annurev.ge.20.120186.000501. [DOI] [PubMed] [Google Scholar]
- 5.Bensi G., Raugei G., Klefenz H., Cortese R. EMBO J. 1985;4:119–126. doi: 10.1002/j.1460-2075.1985.tb02325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smithies O., Connell G.E., Dixon G.H. Nature. 1962;196:232–236. doi: 10.1038/196232a0. [DOI] [PubMed] [Google Scholar]
- 7.Carter K., Worwood M. Int. J. Lab. Hematol. 2007;29:92–110. doi: 10.1111/j.1751-553X.2007.00898.x. [DOI] [PubMed] [Google Scholar]
- 8.Belov, A.P. and Budjakov, J.S., Sudebno-medicinskay ekspertiza, 1963, vol. 1, pp. 28–29.
- 9.McEvoy S.M., Maeda N. J. Biol. Chem. 1988;263:15740–15743. doi: 10.1016/S0021-9258(19)37650-1. [DOI] [PubMed] [Google Scholar]
- 10.Lee P.L., Lee K.Y., Cheng T.M., Chuang H.C., Wu S.M., Feng P.H., Liu W.T., Chen K.Y., Ho S.C. Sci. Rep. 2019;9:1–8. doi: 10.1038/s41598-018-37186-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shih A.W.Y., Mcfarlane A., Verhovsek M. Am. J. Hematol. 2014;89:443–447. doi: 10.1002/ajh.23623. [DOI] [PubMed] [Google Scholar]
- 12.Wicher K.B., Fries E. J. Mol. Evol. 2007;65:373–379. doi: 10.1007/s00239-007-9002-3. [DOI] [PubMed] [Google Scholar]
- 13.Schaer C.A., Owczarek C., Deuel J.W., Schauer S., Baek J.H., Yalamanoglu A., Hardy M.P., Scotney P.D., Schmidt P.M., Pelzing M., Soupourmas P., Buehler P.W., Schaer D.J. BMC Biotechnol. 2018;18:15. doi: 10.1186/s12896-018-0424-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tseng C.F., Lin C.C., Huang H.Y., Liu H.C., Mao S.J.T. Proteomics. 2004;4:2221–2228. doi: 10.1002/pmic.200300787. [DOI] [PubMed] [Google Scholar]
- 15.Mikkat S., Koy C., Ulbrich M., Ringel B., Glocker M.O. Proteomics. 2004;4:3921–3932. doi: 10.1002/pmic.200400825. [DOI] [PubMed] [Google Scholar]
- 16.John H.A., Purdom I.F. Genet. Res. 1987;50:17–21. doi: 10.1017/S0016672300023284. [DOI] [PubMed] [Google Scholar]
- 17.Polticelli F., Bocedi A., Minervini G., Ascenzi P. FEBS J. 2008;275:5648–5656. doi: 10.1111/j.1742-4658.2008.06690.x. [DOI] [PubMed] [Google Scholar]
- 18.Larsson, M., Cheng, T.-M., Chen, C.-Y., and Mao, S.J.T., in Unique Assembly Structure of Human Haptoglobin Phenotypes 1-1, 2-1, and 2-2 and a Predominant Hp 1 Allele Hypothesis. Acute Phase Proteins, INTECH 2013, Chapter 7, pp. 163–179.
- 19.Yang F., Brune J.L., Baldwin W.D., Barnett D.R., Bowman B.H. Proc. Natl. Acad. Sci. USA. 1983;80:5875–5879. doi: 10.1073/pnas.80.19.5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wicher K.B., Fries E. Proc. Natl. Acad. Sci. USA. 2004;101:14390–14395. doi: 10.1073/pnas.0405692101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buehler P.W., Humar R., Schaer D.J. Trends Mol. Med. 2020;26:683–697. doi: 10.1016/j.molmed.2020.02.004. [DOI] [PubMed] [Google Scholar]
- 22.Boettger, L.M., Salem, R.M., Robert, E., Handsaker, R.E., Peloso, G., Kathiresan, S., Hirschhorn, J., and McCarroll, S.A., Nat. Genet., 2016, vol. 48, vol. 4, pp. 359–366. [DOI] [PMC free article] [PubMed]
- 23.Koda Y., Soejima M., Yoshioka N., Kimura H. Am. J. Hum. Genet. 1998;62:245–252. doi: 10.1086/301701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park K.U., Song J., Kim J.Q. J. Clin. Pathol. 2004;57:1094–1095. doi: 10.1136/jcp.2004.017582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurosky A., Barnett D.R., Lee T.-H, Touchstone B., Hay R.E., Arnott M.S., Bowman B.H., Fitch A.W. Proc. Natl. Acad. Sci. USA. 1980;77:3388–3392. doi: 10.1073/pnas.77.6.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alayash A.I., Andersen C.B.F., Moestrup S.K., Bülow L. Trends Biotechnol. 2013;31:2–3. doi: 10.1016/j.tibtech.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 27.Wassler M., Fries E. J. Cell Biol. 1993;123:285–291. doi: 10.1083/jcb.123.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanley J.M., Haugen T.H., Heath E.C. J. Biol. Chem. 1983;258:7858–7869. doi: 10.1016/S0021-9258(18)32258-0. [DOI] [PubMed] [Google Scholar]
- 29.Tripathi A., Lammers K.M., Goldblum S., Shea-Donohue T., Netzel-Arnettb S., Buzza M.S., Antalis T.M., Vogel S.N., Zhao A., Yang S., Arriettac M.-C., Meddingsd J.B., Fasano A. Proc. Natl. Acad. Sci. USA. 2009;106:16799–16704. doi: 10.1073/pnas.0906773106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fasano A. Physiol. Rev. 2011;91:151–175. doi: 10.1152/physrev.00003.2008. [DOI] [PubMed] [Google Scholar]
- 31.Gast M.C.W., van Tinteren H., Bontenbal M., van Hoesel R.Q.G.C.M., Nooij M.A., Rodenhuis S., Span P.N., Tjan-Heijnen V. CG., de Vries E. GE., Harris N., Twisk J.WR., Schellens J. H.M., Beijnen J.H. BMC Cancer. 2008;8:389. doi: 10.1186/1471-2407-8-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clerc F., Reiding K.R., Jansen B.C., Kammeijer G.S.M., Bondt A., Wuhrer M. Glycoconj. J. 2016;33:309–343. doi: 10.1007/s10719-015-9626-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beeri M.S., Lin H.M., Sano M., Ravona-Springer R., Liu X., Bendlin B.B., Gleason C.E., Guerrero-Berroa E., Soleimani L., Launer L.J., Ehrenberg S., Lache O., Seligman Y.K., Levy A.P. JAMA Netw. Open. 2018;1:e184458. doi: 10.1001/jamanetworkopen.2018.4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wobeto V.P. de A., Zaccariotto T.R., Sonati M. de F. Genet. Mol. Biol. 2008;31:602–620. doi: 10.1590/S1415-47572008000400002. [DOI] [Google Scholar]
- 35.Ko D.H., Chang H.E., Kim T.S., Song E.Y., Park K.U., Song J., Han K.S. Biomed. Res. Int. 2013;2013:390630. doi: 10.1155/2013/390630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simpson M., Snell-Bergeon J.K., Kinney G.L., Lache O., Miller-Lotan R., Anbinder Y., Rewers M.J., Levy A.P. Cardiovasc. Diabetol. 2011;10:99. doi: 10.1186/1475-2840-10-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith, J.G. and Gerszten, R.E., Circulation, 2017, vol. 135, vol. 17, pp. 1651–1664. [DOI] [PMC free article] [PubMed]
- 38.Gollapalli K., Ray S., Srivastava R., Renu D., Singh P., Dhali S., Dikshit J.B., Srikanth R., Moiyadi A., Srivastava S. Proteomics. 2012;12:2378–2390. doi: 10.1002/pmic.201200002. [DOI] [PubMed] [Google Scholar]
- 39.Gupta M.B., Seferovic M.D., Liu S., Gratton R.J. Doherty-Kirby, A., Lajoie, G.A., and Han, V.K.M. Clinical Proteomics. 2006;2:169–184. doi: 10.1007/BF02752499. [DOI] [Google Scholar]
- 40.Nirala N.R., Shtenberg G. Biomolecules. 2019;9:12–19. doi: 10.3390/biom9080372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raynes J.G., Eagling S., Mcadam K.P.W.J. Clin. Exp. Immunol. 1991;83:488–491. doi: 10.1111/j.1365-2249.1991.tb05666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pakharukov N.A., Pastushkova L.K., Moshkovskii S.A., Larina I.M. Biochem. Suppl. Ser. B Biomed. Chem. 2011;5:203–212. [Google Scholar]
- 43.Hortin G., Sviridov D., Anderson N.L. Clin. Chem. 2008;54:1608–1616. doi: 10.1373/clinchem.2008.108175. [DOI] [PubMed] [Google Scholar]
- 44.Sadrzadeh S.M.H., Bozorgmehr J. J. Clin. Pathol. 2004;121:S97–104. doi: 10.1309/8GLX5798Y5XHQ0VW. [DOI] [PubMed] [Google Scholar]
- 45.Oh M.K., Park H.J., Lee J.H., Bae H.M., Kim I.S. FEBS Lett. 2015;589:1009–1017. doi: 10.1016/j.febslet.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 46.Di Masi A, De Simone G., Ciaccio C., D’Orso S., Coletta M., Ascenzi P. Mol. Aspects Med. 2020;73:100851. doi: 10.1016/j.mam.2020.100851. [DOI] [PubMed] [Google Scholar]
- 47.McCormick D.J., Atassi M.Z. J. Protein Chem. 1990;9:735–742. doi: 10.1007/BF01024768. [DOI] [PubMed] [Google Scholar]
- 48.Lustbader J.W., Arcoleo J.P., Birken S., Greer J. J. Biol. Chem. 1983;258:1227–1234. doi: 10.1016/S0021-9258(18)33183-1. [DOI] [PubMed] [Google Scholar]
- 49.Bale B.F., Doneen A.L., Vigerust D.J. Front. Cardiovasc. Med. 2018;5:141. doi: 10.3389/fcvm.2018.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Melamed-Frank M., Lache O., Enav B.I., Szafranek T., Levy N.S., Ricklis R.M., Levy A.P. Blood. 2001;8:3693–3698. doi: 10.1182/blood.V98.13.3693. [DOI] [PubMed] [Google Scholar]
- 51.Goldenstein H., Levy N.S., Levy A.P. Pharmacol. Res. 2012;66:1–6. doi: 10.1016/j.phrs.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Levy A.P., Hochberg I., Jablonski K., Resnick H.E., Lee E.T., Best L., Howard B.V. J. Am. Coll. Cardiol. 2002;40:1984–1990. doi: 10.1016/S0735-1097(02)02534-2. [DOI] [PubMed] [Google Scholar]
- 53.Aghaalikhani N., Zamani M., Allameh A., Mashayekhi A., Shadpour P., Mahmoodi M., Rashtchizadeh N. EXCLI J. 2020;19:351–359. doi: 10.17179/excli2019-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaiser M., Thurner EM., Mangge H. Sci. Rep. 2020;10:13117.. doi: 10.1038/s41598-020-69333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tan S.H., Yougbaré S., Chu HL., Kuo T.R., Cheng T.M. Polymers. 2020;12:2242. doi: 10.3390/polym12102242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.MacKellar M., Vigerust D.J. Clin. Diabetes. 2016;34:148–157. doi: 10.2337/diaclin.34.3.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ascenzi P., Bocedi A., Visca P., Altruda F., Tolosano E., Beringhelli T., Fasano M. IUBMB Life. 2005;57:749–759. doi: 10.1080/15216540500380871. [DOI] [PubMed] [Google Scholar]
- 58.Lipiski M., Deuel J.W., Baek J.H., Engelsberger W.R., Buehler P.W., Schaer D.J. Antioxidants Redox Signal. 2013;19:1619–1633. doi: 10.1089/ars.2012.5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheng T.M., Pan J.P., Lai S.T., Kao L.P., Lin H.H., Mao S.J.T. Clin. Biochem. 2007;40:1045–1056. doi: 10.1016/j.clinbiochem.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 60.Bamm V. V., Tsemakhovich V.A., Shaklai M., Shaklai N. Biochemistry. 2004;43:3899–3906. doi: 10.1021/bi0362626. [DOI] [PubMed] [Google Scholar]
- 61.Schaer D.J., Buehler P.W., Alayash A.I., Belcher J.D., Vercellotti G.M. Blood. 2013;121:1276–1284. doi: 10.1182/blood-2012-11-451229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Kleijn D.P.V., Smeets M.B., Kemmeren P.P.C.W., Lim S.K., van Middelaar B.J., Velema E., Schone-veld A., Pasterkamp G., Borst C. FASEB J. 2002;16:1123–1125. doi: 10.1096/fj.02-0019fje. [DOI] [PubMed] [Google Scholar]
- 63.Nantasenamat C., Prachayasittikul V., Bulow L. PLoS One. 2013;8:e62996. doi: 10.1371/journal.pone.0062996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lim S.K. Redox Rep. 2001;6:375–378. doi: 10.1179/135100001101536571. [DOI] [PubMed] [Google Scholar]
- 65.Nielsen M.J., Moestrup S.K. Blood. 2009;108:2846–2849. doi: 10.1182/blood-2006-05-022327. [DOI] [PubMed] [Google Scholar]
- 66.Tolosano E., Fagoonee S., Hirsch E., Berger F.G., Baumann H., Silengo L., Altruda F. Blood. 2002;100:4201–4205. doi: 10.1182/blood-2002-04-1270. [DOI] [PubMed] [Google Scholar]
- 67.Arredouani M., Matthijs P., Van Hoeyveld E., Kasran A., Baumann H., Ceuppens J.L., Stevens E. Immunology. 2003;108:144–151. doi: 10.1046/j.1365-2567.2003.01569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Asleh R., Marsh S., Shilkrut M., Binah O., Guetta J., Lejbkowicz F., Enav B., Shehadeh N., Kanter Y., Lache O., Cohen O., Levy N.S., Levy A.P. Circ. Res. 2003;92:1193–1200. doi: 10.1161/01.RES.0000076889.23082.F1. [DOI] [PubMed] [Google Scholar]
- 69.Cahill L.E., Jensen M.K., Chiuve S.E., Shalom H., Pai J.K., Flint A.J., Mukamal K.J., Rexrode K.M., Levy A.P., Rimm E.B. J. Am. Coll. Cardiol. 2015;66:1791–1799. doi: 10.1016/j.jacc.2015.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cahill L.E., Levy A.P., Chiuve S.E., Jensen M.K., Wang H., Shara N.M., Blum S., Howard B.V., Pa J.K., Mukamal K.J., Rexrode K.M., Rimm E.B. J. Am. Coll. Cardiol. 2013;61:728–737. doi: 10.1016/j.jacc.2012.09.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Asleh, R., Guetta, J., Kalet-Litman, S., Miller-Lotan, R., and Levy, A.P., Circ. Res., 2005, vol. 96, vol. 435–441. [DOI] [PubMed]
- 72.Eaton J.W., Brandt P., Mahoney J.R., Lee J.T. Science. 1982;215:691–693. doi: 10.1126/science.7036344. [DOI] [PubMed] [Google Scholar]
- 73.Yang F., Friedrichs W.E., Navarijo-Ashbaugh A.L., DeGraffenried L.A., Bowman B.H., Coalson J.J. Lab. Investig. 1995;73:433–440. [PubMed] [Google Scholar]
- 74.Langlois M.R., Delanghe J.R. Clin. Chem. 1996;42:1589–1600. doi: 10.1093/clinchem/42.10.1589. [DOI] [PubMed] [Google Scholar]
- 75.Sertório J.T., Lacchini R., Amaral L.M., Palei A.C.T., Cavalli R.C., Sandrim V.C., Duarte G., Tanus-Santos J.E. J. Hum. Hypertens. 2013;27:349–354. doi: 10.1038/jhh.2012.57. [DOI] [PubMed] [Google Scholar]
- 76.Cid M.C., Grant D.S., Hoffman G.S., Auerbach R., Fauci A.S., Kleinman H.K. J. Clin. Invest. 1993;91:977–985. doi: 10.1172/JCI116319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pavlíček Z., Ettrich R. Collect. Czechoslov. Chem. Commun. 1999;4:717–725. doi: 10.1135/cccc19990717. [DOI] [Google Scholar]
- 78.Vasilevsky I.V. Mezhdunarodnye obzori klinicheskaya praktika i zdorovje. 2017. [Google Scholar]
- 79.Guerranti R., Bertocci E., Fioravanti A., Papakostas P. Int. J. Immunopathol. Pharmacol. 2010;23:901–909. doi: 10.1177/039463201002300326. [DOI] [PubMed] [Google Scholar]
- 80.Cavalli-Sforza L.L., Menozzi P., Piazza A. The History and Geography of Human Genes. Princeton: Princeton University Press; 1994. [Google Scholar]
- 81.Langlois M., Delanghe J. Clin. Chem. 1996;42:1589–6008. doi: 10.1093/clinchem/42.10.1589. [DOI] [PubMed] [Google Scholar]
- 82.Delanghe J.R., Marijn M., Speeckaert M.M., De Buyzere M.L. Clin. Chem. Lab. Med. 2020;58:1125–1126. doi: 10.1515/cclm-2020-0425. [DOI] [PubMed] [Google Scholar]
- 83.Nielsen M.J., Petersen S.V., Jacobsen C., Oxvig C., Rees D., Møller H.J., Moestrup S.K. Blood. 2006;108:2846–2849. doi: 10.1182/blood-2006-05-022327. [DOI] [PubMed] [Google Scholar]
- 84.Peacock A.C. J. Natl. Cancer Inst. 1966;36:631–639. doi: 10.1093/jnci/36.4.631. [DOI] [PubMed] [Google Scholar]
- 85.Nikolenko, O.V., Sudebno-medicinskay ekspertiza, 1977, vol. 3, pp. 45–47.
- 86.Pompach P., Brnakova Z., Sanda M., Wu J., Edwards N., Goldman R. Mol. Cell. Proteomics. 2013;12:1281–1293. doi: 10.1074/mcp.M112.023259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen C.B, Su Y.C., Huang T.T., Ho H.C., Chang Y.-T., Tung Y.-T., Lee W.C. Clin. Chim. Acta. 2008;398:48–52. doi: 10.1016/j.cca.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 88.Sanchez J.-C., Appel R.D., Golaz O., Pasquali C., Ravier F., Bairoch D F., Hochstrasse D.F. Electrophoresis. 1995;16:1131–1151. doi: 10.1002/elps.11501601190. [DOI] [PubMed] [Google Scholar]
- 89.Bahk Y.Y., Na B.K. Cho, S.H., Kim, J.Y. Lim, K.-J., and Tong-Soo Kim, T-S. Korean J. Parasitol. 2010;48:203–211. doi: 10.3347/kjp.2010.48.3.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ma H.T., Sriyam S., Sinchaikul S., Tsai H.Y., Phutrakul S., Chen S.-T. J. Proteomics Bioinform. 2013;6:187–196. [Google Scholar]
- 91.Okano T., Seike M., Kuribayashi H., Soeno C., Ishii T., Kida K., Gemma A. Int. J. Oncol. 2016;48:945–952. doi: 10.3892/ijo.2016.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Edelman M.J., Hodgson L., Rosenblatt P.Y., Christenson R.H., Vokes E.E., Wang X., Kratzke R.P.Y. J.Thorac. Oncol. 2012;7:649–654. doi: 10.1097/JTO.0b013e31824a8db0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhao C., Annamalai L., Guo C., Kothandaraman N., Koh S.C.L., Zhang H., Arijit Biswas A., Choolani M. Neoplasia. 2007;9:1–7. doi: 10.1593/neo.06619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Garibay-Cerdenares O.L., Hernández-Ramírez V.I., Osorio-Trujillo J.C., Hernández-Ortíz M., Gallardo-Rincón D., de León D.C., Encarnación-Guevara S., Villegas-Pineda J.C., Talamás-Rohana P. J. Ovarian Res. 2014;7:27. doi: 10.1186/1757-2215-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ye B., Cramer D.W., Skates S.J., Gygi S.P., Pratomo V., Fu L., Horick N.K., Licklider L.J., Schorge J.O., Berkowitz R.S., Mok S.C. Clin. Cancer Res. 2003;9:2904–2911. [PubMed] [Google Scholar]
- 96.Chen J., Cheuk I.W., Siu M.T., Yang W., Cheng A.S., Shin V.Y., Kwong A. Am J. Cancer Res. 2020;10:2865–2877. [PMC free article] [PubMed] [Google Scholar]
- 97.Bakrim N.M., Mohd S.A.N.S., Talib A.N., Ab Rahman J., Abdullah A. Malays J. Med. Sci. 2020;27:64–76. doi: 10.21315/mjms2020.27.2.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chiappin S., Antonelli G., Gatti R., De Palo E.F. Clin. Chim. Acta. 2007;383:30–40. doi: 10.1016/j.cca.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 99.Zhang X.L., Wu Z.Z., Xu Y., Wang J.G., Wang Y.Q., Cao M.Q., Wang C.H. Open Chem. 2020;18:918–926. doi: 10.1515/chem-2020-0048. [DOI] [Google Scholar]
- 100.Sun L., Hu S., Yu L., Guo C., Sun L., Yang Z., Qi J., Ran Y. Int. J. Cancer. 2016;138:2724–2731. doi: 10.1002/ijc.29993. [DOI] [PubMed] [Google Scholar]
- 101.Kumar D.M., Thota B., Shinde S.V., Prasanna K.V., Hegde A.S., Arivazhagan A., Chandramouli B.A., Santosh V., Somasundaram K. J. Proteome Res. 2010;9:5557–5567. doi: 10.1021/pr1001737. [DOI] [PubMed] [Google Scholar]
- 102.Petushkova N.A., Zgoda V.G., Pyatnitskiy M.A., Larina O.V., Teryaeva N.B., Potapov A.A., Lisitsa A.V. PLoS One. 2017;12:e0177427. doi: 10.1371/journal.pone.0177427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Okuyama N., Ide Y., Nakano M., Nakagawa T., Yamanaka K., Moriwaki K., Murata K., Ohigashi H., Yokoyama S., Eguchi H., Ishikawa O., Ito T., Kato M., Kasahara A., Kawano S., Gu J., Taniguchi N., Miyoshi E. Int. J. Cancer. 2006;118:2803–2808. doi: 10.1002/ijc.21728. [DOI] [PubMed] [Google Scholar]
- 104.Ang I.L., Poon T.C.W., Lai P.B.S., Chan A.T.C., Ngai S.-M., Hui A.Y., Johnson P.J., Sung J.J.Y. J. Proteome Res. 2006;5:2691–2700. doi: 10.1021/pr060109r. [DOI] [PubMed] [Google Scholar]
- 105.Wu D., Struwe W.B., Harvey D.J., Fergu-son M.A.J., Robinson C.V. Proc. Natl. Acad. Sci. USA. 2018;115:8763–8768. doi: 10.1073/pnas.1807439115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Park S.Y., Lee S.H., Kawasaki N., Itoh S., Kang K., Ryu S.H., Hashii N., Kim J.-M., Kim J.-Y., Kim J.H. Int. J. Cancer. 2012;130:2366–2376. doi: 10.1002/ijc.26288. [DOI] [PubMed] [Google Scholar]
- 107.Hamrita B., Chahed K., Trimeche M., Guillier C.L., Hammann P., Chaïeb A., Korbi S., Chouchane L. Clin. Chim. Acta. 2009;404:111–118. doi: 10.1016/j.cca.2009.03.033. [DOI] [PubMed] [Google Scholar]
- 108.Nakano, M., Nakagawa, T., Ito, T., Kitada, T., Hijioka, T., Kasahara, A., Tajiri, M., Wada, Y., Taniguchi, N., and Miyoshi, E., Int. J. Cancer, 2008, vol. 122, vol. 10, pp. 2301–2309. [DOI] [PubMed]
- 109.Hoagland L.F.M., Campa M.J., Gottlin E.B., Herndon J.E., Patz E.F. Cancer. 2007;110:2260–2268. doi: 10.1002/cncr.23049. [DOI] [PubMed] [Google Scholar]
- 110.Lee S.H., Jeong S., Lee J., Yeo I.S., Oh M.J., Kim U., Kim S., Kim S.H., Park S.-Y., Kim J.-H., Park S.H., Kim J.H., An H.J. Mol. Biosyst. 2016;12:3611–3621. doi: 10.1039/C6MB00559D. [DOI] [PubMed] [Google Scholar]
- 111.Kim J.H., Lee S.H., Choi S., Kim U., Yeo I.S., Kim S.H., Oh M.J., Moon H., Lee J., Jeong S., Choi M.G., Lee J.H., Sohn T.S., Bae J.M., Kim S., Min Y.W., Lee H., Lee J.H., Rhee P.-L., Kim J.J., Lee S.J., Kim S.T., Lee J., Park S.H., Park J.O., Park Y.S., Lim H.Y., Won Ki Kang W.K., An H.J., Kim J.H. Oncotarget. 2017;8:11094–11104. doi: 10.18632/oncotarget.14362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Miyoshi E., Moriwaki K., Terao N., Tan C.C., Te-rao M., Nakagawa T., Matsumoto H., Shinzaki S., Kamada Y. Biomolecules. 2012;2:34–45. doi: 10.3390/biom2010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Maresca B., Cigliano L., Spagnuolo M.S., Piaz F.D., Corsaro M.M., Balato N., Nino M., Balato A., Ayala F., Abrescia P. PLoS One. 2012;7:e52040. doi: 10.1371/journal.pone.0052040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nishino K., Koda S., Kataoka N., Takamatsu S., Nakano M., Ikeda S., Kamamatsu Y., Morishita R., Moriwaki K., Eguchi H., Yamamoto E., Kikkawa F., Tomita Y., Kamada Y., Miyoshi E. Oncotarget. 2018;9:12732–12744. doi: 10.18632/oncotarget.24332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Morishita K., Maki Y, Takamatsu S., Ito N., Koda S., Kei Motooka K., Kamada Y., Kajihara Y., Miyoshi E. Analytical Biochemistry. 2020;593:113588. doi: 10.1016/j.ab.2020.113588. [DOI] [PubMed] [Google Scholar]
- 116.Zhang S., Shang S., Li W., Qin X., Liu Y. Glycobiology. 2016;26:684–692. doi: 10.1093/glycob/cww016. [DOI] [PubMed] [Google Scholar]
- 117.Tamara S., Franc V., Heck A.J.R. Proc. Natl. Acad. Sci. USA. 2020;117:15554–15564. doi: 10.1073/pnas.2002483117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dotz V., Wuhrer M. FEBS Lett. 2019;593:2966–2976. doi: 10.1002/1873-3468.13598. [DOI] [PubMed] [Google Scholar]


