FIGURE 4.
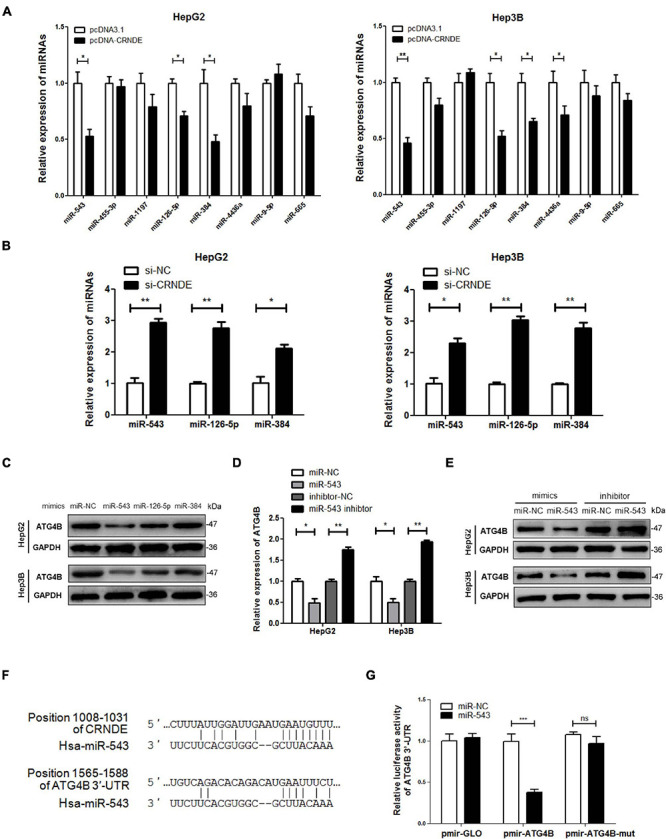
miR-543 downregulates ATG4B by targeting its 3′-UTR. (A,B) HepG2 and Hep3B cells were transfected with pcDNA-CRNDE (or pcDNA3.1) (A) or si-CRNDE (or si-NC) (B) for 24 h, then the indicated miRNAs were analyzed by qPCR. (C) HepG2 and Hep3B cells were separately transfected with the mimics of the three candidate miRNAs (or miR-NC) for 24 h, then ATG4B protein level was measured by Western blot. (D,E) HepG2 and Hep3B cells were transfected with miR-543 mimics (or miR-NC) or inhibitor (or inhibitor-NC) for 24 h, then the levels of ATG4B mRNA and protein were separately detected by qPCR (D) and Western blot (E). (F) The predicted binding sites of miR-543 in CRNDE and 3′-UTR of ATG4B mRNA. (G) HepG2 cells were co-transfected with the luciferase reporter plasmid pmir-ATG4B (or pmir-ATG4B-mut, or pmirGLO) and miR-543 mimics (or miR-NC) for 24 h. Then the luciferase activities were determined by dual-luciferase reporter assay. The activity of firefly luciferase was normalized against that of rennilla luciferase, and the result was shown as relative luciferase activity. miR-NC: control mimics; inhibitor-NC: control inhibitor; pmir-ATG4B: reporter vector of 3′-UTR of ATG4B mRNA; pmir-ATG4B-mut: reporter vector of mutant 3′-UTR of ATG4B mRNA; pmir-GLO: pmir-GLO vector; pcDNA-CRNDE, pcDNA3.1, si-CRNDE and si-NC were the same as the description in Figure 1; ns, no significance; *P < 0.05; **P < 0.01; ***P < 0.001.
