Abstract
Background
The gain/amplification CKS1B gene at chromosome region 1q21 (1q+) is one of the most common genetic aberrations in multiple myeloma (MM). Amplification of CKS1B is frequently associated with the deletion of CDKN2C gene at chromosome region 1p32 (1p-), which is also associated with inferior outcomes.
Methods
In this retrospective study, we evaluated the outcomes of patients with 1q+ and/or 1p- after high-dose therapy and auto-HCT. From January 2006 to December 2015, 1491 newly diagnosed MM patients underwent upfront high-dose therapy and auto-HCT at our institution. Out of those, 899 had the fluorescent in situ hybridization (FISH) data available. FISH was performed at diagnosis and before the start of induction in 686 (76%) patients, and after the initiation of induction therapy in 213 (24%) patients. We identified 100 patients with 1q+ and/or 1p- by FISH from the cohort of 899 patients. A control group (N=287) with diploid cytogenetics and normal FISH panel was selected from the same cohort. From the above two cohorts, using a propensity-score matched analysis, we identified matched controls for 85 of the 100 patients with 1q+/1p−. Patients were matched for age at auto-HCT, sex, international staging system stage, induction regimen, creatinine level, disease status at auto-HCT, conditioning regimen and maintenance therapy.
Results
Sixty-seven (79%), 4 (5%) and 14 (16%) patients had 1q+, 1p- or both 1q+ and 1p-, respectively. There was no significant difference in induction therapy, preparative regimen, or maintenance therapy between the 1q+/1p− and control groups. The median follow-up time for all patients was 29.2 months (range: 0.29 −84.96). The cumulative incidence of 100-day non-relapse mortality was 1.2% and 0% for the 1q+/1p− and the control groups, respectively. Forty-two patients (50%) in the 1q+/1p− group achieved complete response compared to 40 patients (47%) in the control group. The estimated 3-year progression-free survival (PFS) and overall survival (OS) rates for the 1q+/1p− and the control groups were 41% and 79%, and 56% and 86%, respectively. Patients in the 1q+/1p− group experienced significant increased risk of progression or death compared with the control group (HR 2.21, CI 1.18–4.16, P=0.014). No significant association between OS in the two groups were observed. The outcome of the 1q+/1p− alone (with no additional high-risk cytogenetics) and the propensity-score matched control groups was also compared. Median PFS for the 1q+/1p− alone subgroup was 26.6 months, compared to 38.8 months for the control group (HR 1.9, CI 0.9–4.08, P=0.09). The median OS had not reached for the 1q+1/p- alone subgroup and was 81.1 months for the control group (HR 1.25, CI 0.3–4.6, P=0.73)
Conclusion
1q+/1p− abnormalities with amplification of CKS1B and deletion of CDKN2C genes were associated with shorter PFS when compared to a propensity-score matched group of patients with diploid cytogenetics and normal a FISH panel. The outcomes of 1q+1/p- MM patients have improved with the use of more effective induction, conditioning, and maintenance therapy compared to historical controls, but they still need more effective therapeutic approaches to fully overcome the negative impact of 1q+1/p-.
Keywords: Multiple myeloma, CKS1B, CDKN2C, autologous stem cell transplantation
INTRODUCTION
Multiple myeloma (MM), the second most common hematological malignancy, is a clonal plasma cell disorder with a heterogeneous disease course. There is a large variability in the outcome 1–3. A number of factors affect the outcome of MM patients, including cytogenetic abnormalities and the ISS/R-ISS stage 4–6. The cytogenetic abnormalities in MM can be divided into primary and secondary abnormalities 7. The two primary cytogenetic abnormalities are: a) trisomies, typically of the chromosomes 3, 5, 9, 11, 15, 19, and 21, b) translocations involving the immunoglobulin heavy chain (IgH), where an oncogene from another chromosome is translocated to the IgH region of 14q32. Monosomies (eg monosomy 14) can also sometimes be primary abnormalities. Secondary cytogenetic abnormalities that drive the disease progression are associated with evolution of monoclonal gammopathy of undetermined significance and smoldering myeloma to symptomatic MM, and frequently involve chromosome 13, deletion of chromosome 17p, duplication of chromosome 1q21 and deletion of 1p. The gain/amplification of CKS1B gene at chromosome region 1q21 (1q+) is one of the most common secondary genetic abnormalities in MM, and plays a critical role in cell cycle progression and MM cell survival.
CKS1B, an essential protein for cell growth and division, is a member of cyclin kinase subunit 1 protein family and is expressed universally in the bone marrow and various other tissues. CKS1B associates with p27kip1-Cdk/cyclin complex and acts as a cofactor for Skp2-dependent ubiquitination of p27 8,9. An amplified CKS1B results in greater degradation of p27, activation of the Cdk/cyclin complex, and cell cycle up-regulation by promoting G1/S transition 10.
The prognostic role of CKS1B gain/amplification in MM remains controversial. Some groups, including ours, have shown that the presence of 1q+ is associated with poor survival outcomes in MM 10–13. In contrast, other groups failed to show 1q+ as an independent prognostic marker and concluded that the concurrent presence of other known high-risk chromosomal abnormalities was associated with worse outcomes 14.
Amplification of CKS1B is frequently associated with the deletion of CDKN2C gene at chromosome 1p32.3 (1p-) locus. Deletion of CDKN2C, a tumor suppressor gene, leads to deregulation of G1/S transition, resulting in an increased proliferative rate of plasma cells in MM patients 15. Many studies, including ours, have shown the deletion of 1p as a high-risk genetic abnormality associated with poor prognosis in MM 16–18.
The negative impact of some of the high-risk genetic abnormalities is abated with the use of novel proteasome inhibitors, such as bortezomib, in the induction regimen19. However, the outcomes of both 1q+/1p− continue to be poor despite the use of novel induction therapies. There is limited data on the impact of 1q+/1p− in newly diagnosed MM patients after high-dose therapy and autologous hematopoietic cell transplantation (auto-HCT). As both 1q+ and 1p- are secondary cytogenetic abnormalities7 and most of the high risk genes mapped on chromosome 1 either resided on 1q or 1p20, our study focused on evaluating the outcomes of newly diagnosed MM patients with 1q+ and/or 1p- after high-dose therapy and auto-HCT, and compared their outcomes to a propensity-score matched control group of patients who had diploid cytogenetics and a normal MM fluorescent in situ hybridization (FISH) panel.
METHOD
From January 2006 to December 2015, 1491 newly diagnosed MM patients underwent upfront high-dose therapy and auto-HCT at our institution. Out of those, 899 had FISH data available. The FISH studies included testing for TP53/CEN17,MYEOV/CCND1-IGH/t(11;14),FGFR3-IGH/t(4;14),MAF-IGH/t(14;16),RB1 and CDKN2C/CKS1B. FISH was performed at diagnosis and before the start of induction in 686 (76%) patients, and after the initiation of induction therapy in 213 (24%) patients. We identified 100 patients with 1q+ (CKS1B) and/or 1p- (CDKN2C) from that cohort, who had these abnormalities detected on FISH at any time before auto-HCT. Most of these samples were not CD138 enriched as we did not routinely perform CD138 enrichment up until recently. The 95% (P<0.05) confidence limit of the CDKN2C/CKS1B probes established on twenty normal samples using the Beta Inverse Method of calculation at our cytogenetics laboratory for 1p32.3/CDKN2C deletion in interphase cells was 0.0–6.8, and for 1q-21/CKS1B gain/amplification for 3 signals was 0.0–7.9%, and for 4 or more signals was 0.0–4.4%. A control group (N=287) with diploid cytogenetics and normal MM FISH panel was selected from the same cohort. From the above two cohorts, using a propensity-score matched analysis, we identified matched controls for 85 of the 100 patients with 1q+/1p−. In the 1q+/1p− and the control groups, FISH was performed at diagnosis and before the start of induction in 64 (75%) and 57 (67%) patients, respectively.
Patients were defined as having high-risk MM if they had 17p, t(4;14), t(14;16), t(14;20), chromosomal 1 abnormalities (CKS1B gain/amplification and CDKN2C deletion) on FISH studies. To evaluate the effect of copy number variation on outcomes after auto-HCT, the gain/amplification of 1q+ was divided into two categories21: 1) 3 copies: cells with at least 4 or more copies of 1q+ were seen in fewer than 20% of the clonal plasma cells; and 2) ≥ 4 copies: cells with 4 or more copies of 1q+ were seen in more than 20% of the clonal plasma cells.
Response and Outcome
The International Myeloma Working Group (IMWG) uniform response criteria were used to calculate the response to induction treatment and auto-HCT22,23.
The overall response rate (ORR) was assessed at 3 months after auto-HCT and included patients who achieved a complete response (CR), very good partial response (VGPR) and partial response (PR).
Statistical Methods
To correct for biases in the data, the nearest-neighbor matching method was employed, using a caliper of 0.25, where a control patient within the caliper for a treated patient was randomly selected as the match for that treated patient. Variables used for matching were age at auto-HCT, sex, international staging system stage, induction regimen, creatinine level, disease status at auto-HCT, conditioning regimen and maintenance therapy. One-hundred patients with 1q+ and/or 1p- by FISH were identified in this analysis. Of those, 9 patients had missing information for at least one measure that was included in the propensity score matching (100–9=91). The matching method used for the cases was nearest neighbor matching within a specified caliper distance (0.25), where the absolute difference in the propensity scores between matched subjects (i.e., the controls) and the 1q+1/p- were required to be less than 0.25. Six cases did not have controls that met the caliper criteria (91–6=85). Therefore, 85 cases and matched-controls were reported in this paper. Cox proportional hazards regression models stratifying on the matched pairs were used to estimate the association between the matched group and survival outcomes (overall survival [OS] and progression-free survival [PFS]). PFS was calculated from the date of auto-HCT to the date of progression or death from any cause and OS was calculated from the date of auto-HCT to the date of death from any cause. The Kaplan-Meier method was used to estimate OS and PFS and group differences were evaluated by the stratified log-rank test. The log-rank test was used to test the null hypothesis that there was no difference between populations in the probability of an event at any time point. The analysis was based on the times of events. Three assumptions were associated with the log-rank test: i) at any time patients who were censored have the same survival prospects as those who continue to be followed (i.e., censoring was unrelated to prognosis); ii) the survival probabilities were the same for subjects recruited early and late in the study; and iii) the event happens at the time specified. The cumulative incidence of non-relapse mortality (NRM) was determined using the competing risks method. The competing risk included was disease progression and patients who were still alive at the last follow-up date were censored. Group differences in NRM were assessed by Gray’s test, stratified by matched group. The Wilcoxon signed-rank test and conditional logistic regression models were conducted to assess the association between prognostic risk factors and 1q+/1p−/control groups as appropriate. No adjustments for multiple testing were performed. With the exception of matching, all statistical analyses were performed using SAS 9.4 for Windows (copyright © 2011 by SAS Institute Inc., Cary, NC). All statistical tests used a significance level of 5%. Nearest-neighbor matching was carried out using the MatchIt package in R.
RESULTS
Patient Characteristics
A total of 100 newly diagnosed MM patients out of the 899 patients with available FISH data (11%) with 1q+/1p− (80 pts with 1q+ only, 4 pts with 1p- only, and 16 pts with both 1q+ and 1p) underwent high dose therapy and auto-HCT between 2006–2015 at our center. Using propensity-score matched analysis, we were able to identify a matched control for the 85 patients with 1q+/1p−. The baseline characteristics of the 1q+/1p− and control groups are summarized in Table 1. Sixty-seven (79%), 4 (5%) and 14 (16%) patients had 1q+, 1p-, or both 1q+ and 1p-, respectively.
Table 1:
Baseline characteristics for matched cohort of 1q+/1p− (N=85) and control (N=85) groups
| Matched factors | 1q+/1p− | Control | p-Value* | |
|---|---|---|---|---|
| (N=85) | (N=85) | |||
| Age at transplantation | N, Median | 85, 61.0 | 85, 61.0 | 0.47 |
| Age (Years) | Range | 34.0–77.0 | 44.0–79.0 | |
| Creatinine | N, Median | 85, 1.06 | 85, 1.01 | 0.98 |
| Range | 0.5 – 11.8 | 0.5 – 17.9 | ||
| Sex, n (%) | Female | 35 (41) | 35 (41) | 1.00 |
| Male | 50 (59) | 50 (59) | ||
| ISS Stage, n (%) | I | 30 (35) | 32 (38) | 0.94 |
| II | 29 (34) | 28 (33) | ||
| III | 26 (31) | 25 (29) | ||
| Disease status at auto-HCT, n (%) | PD | 3 (4) | 3 (4) | 0.99 |
| SD | 2 (2) | 2 (2) | ||
| PR | 30 (35) | 32 (38) | ||
| VGPR | 39 (46) | 36 (42) | ||
| CR | 11 (13) | 12 (14) | ||
| Conditioning regimen, n (%) | BuMel | 11 (13) | 13 (15) | 0.64 |
| Melphalan | 74 (87) | 72 (85) | ||
| Induction, n (%) | VCD | 16 (19) | 20 (24) | 0.93 |
| VD | 6 (7) | 5 (6) | ||
| PI + IMiD | 51 (60) | 47 (55) | ||
| IMiD | 1 (1) | 3 (3) | ||
| Others | 11 (13) | 10 (12) | ||
| Maintenance, n (%) | PI+IMiD | 10 (12) | 8 (9) | 0.85 |
| Bortezomib | 11 (13) | 8 (9) | ||
| Lenalidomide+ Elotuzumab | 3 (4) | 4 (5) | ||
| IMiD | 39 (46) | 42 (49) | ||
| None | 22 (26) | 23 (27) | ||
Abbreviations: BuMel, Busulfan-melphalan; CR, complete remission; IMiD, immunomodulatory drug; ISS, International Staging System; PD, progressive disease; PI proteasome inhibitor; PR, partial response; SD, stable disease; VCD, Bortezomib cyclophosphamide dexamethasone; VD, Bortezomib Dexamethasone; VGPR, very good partial response.
Sixty-seven (79%) patients in both the 1q+/1p− and control groups received a triplet-based induction regimen involving a combination of a proteasome inhibitor (PI), dexamethasone and an immunomodulatory drug (IMiD) or cyclophosphamide. ORR (CR, VGPR, PR) in both 1q+/1p− and the control group at the time of auto-HCT was 94% (80/85). Melphalan alone was used as preparative regimen in 74 patients (87%) in the 1q+/1p− group and in 72 patients (85%) in the control group.
Engraftment and Non-relapse Mortality
The median time to neutrophil engraftment was 11 days in both the 1q+/1p− (range, 10 to 14, n=84) and the control groups (range, 8 to 16, n=85). The cumulative incidence of 100-day non-relapse mortality (NRM) was 1.2% and 0% for the 1q+/1p− and the control groups, respectively (P=0.32).
Response
Forty-two patients out of 84 patients with information (50%) in the 1q+/1p− group achieved a CR compared to 40 patients (47%) in the control group. Thirty-one patients (37%) in the 1q+/1p− group, and 35 (41%) in the control group achieved a VGPR. Nine patients (11%) in the 1q+/1p− group and 10 patients (12%) in the control group achieved a PR. The ORR for the 1q+/1p−and the control group was 98% and 100%, respectively (P=0.25).
Relapse and Survival
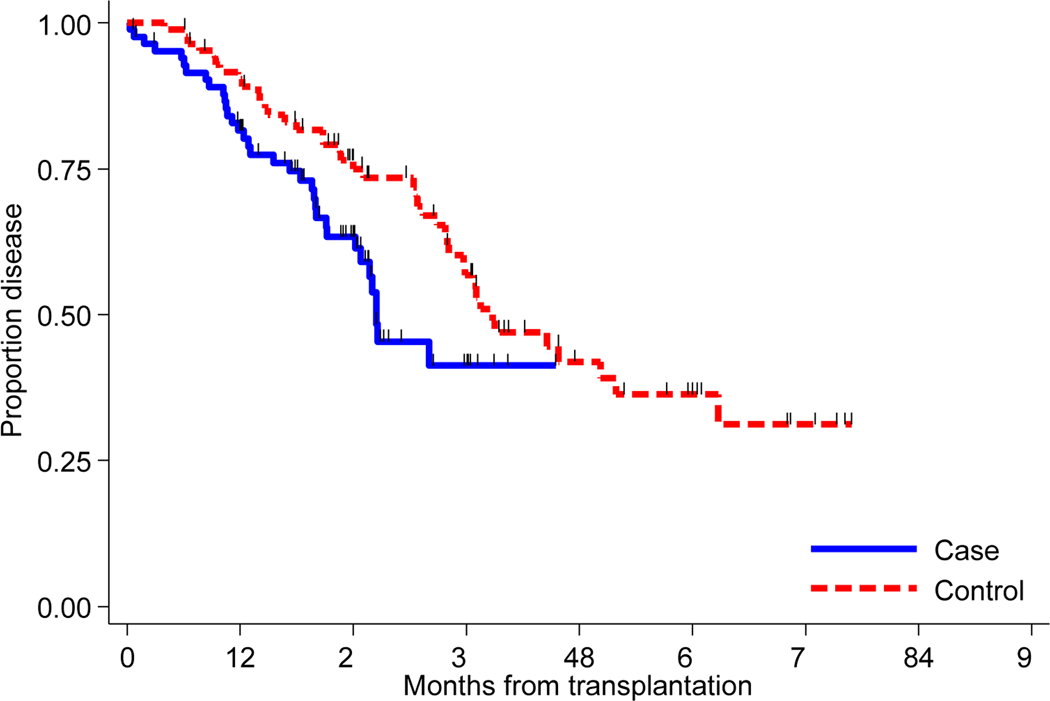
The median follow-up time for all patients was 29.2 months (range: 0.3 −85.0). Out of 85 1q+1/p- patients, 32 progressed. FISH was available at relapse in 14 of the 32 patients. Of those 14 patients, 10 patients had 1q+ before auto HCT and 8 of them continued to have of 1q+ at relapse; 1 patient with 1p- had both 1p- and 1q+ at relapse; 3 patients had both 1q+ and 1p-, one of them remained positive for 1q+ and other two were negative for both 1q+ and 1p-. Median PFS for the 1q+/1p− and the control group was 26.5 months and 38.8 months, respectively. The estimated 3-year PFS rates for the 1q+/1p− and the control group were 41% and 56%, respectively (Figure 1A). In addition, the 1q+/1p− patients experienced increased risk of progression or death compared with the control group (HR 2.21, CI 1.18–4.16, P=0.014).
Figure 1A:
Kaplan-Meier curves of progression free survival for matched cohort of 1q+/1p− (N= 85) and controls (N=85).
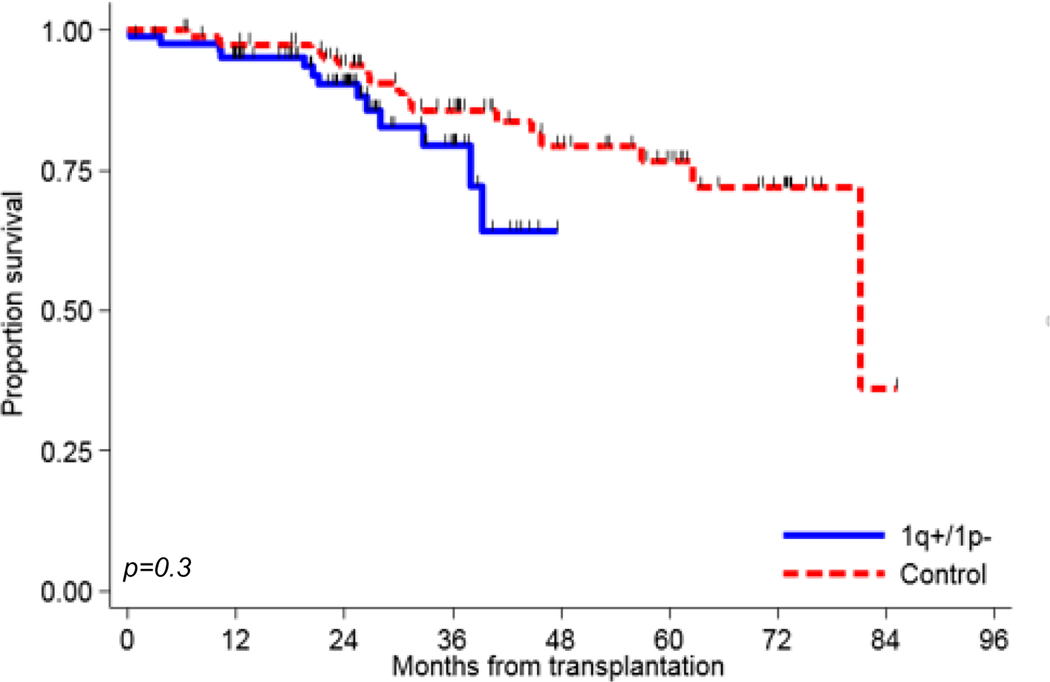
Median OS had not been reached for the 1q+/1p− group, and was 81.1 months for the control group. The 3-year OS rates for the 1q+/1p− and the control group were 79% and 86%, respectively (Figure 1B). No significant association between OS in the two group of patients was noted (HR 1.57, CI 0.61–4.05, P=0.35).
Figure 1B:
Kaplan-Meier curves of overall survival for matched cohort of 1q+/1p− (N= 85) and controls (N=85).
Additional analyses
We performed additional analyses on all 100 patients who had either 1q+ or 1p-. Thirty three (33%) patients with 1q+/1p− had additional high risk cytogenetic abnormalities (17p, t(4;14), t(14;16)) on the FISH panel. None of 1q+/1p− patients had t(14;20). These additional high-risk cryptogenic abnormalities are summarized in Table 2. We compared the outcome of the 1q+/1p− alone (with no additional high-risk cytogenetics) and the propensity-score matched control groups. Median PFS for the 1q+/1p− alone subgroup was 26.6 months, compared to 38.8 months for the control group (HR 1.9, CI 0.9–4.08, P=0.09). The median OS had not reached for the 1q+1/p- alone subgroup and was 81.1 months for the control group (HR 1.25, CI 0.3–4.6, P=0.73) (Table 3)
Table 2:
Additional high risk abnormalities detected on FISH for all 1q+/1p− patients (N=100)
| High risk on FISH | 17p | t(14;16) | t(4;14) |
|---|---|---|---|
| N (%) | 17 (17%) | 8 (8%) | 13 (13%) |
Abbreviation: FISH: fluorescence in situ hybridization.
Table 3:
PFS and OS of matched cohort of 1q+/1p− alone (without any additional high risk cytogenetics) and matched (N=58) controls.
| Variable | Total no. of pts | Median months | HR | HR | HR | P-value |
|---|---|---|---|---|---|---|
| Lower CI | Upper CI | |||||
| PFS | ||||||
| 1q+/1p− alone | 58 | 26.6 | 1.9 | 0.9 | 4.08 | 0.09 |
| Control | 58 | 38.8 | ||||
| OS | ||||||
| 1q+/1p− alone | 58 | NR | 1.25 | 0.3 | 4.6 | 0.73 |
| Control | 58 | 81.1 |
Abbreviations: CI, confidence interval; HR, hazards ratio; NR, not reached; OS, overall survival; PFS, progression- free survival; Ref, reference group
Note: High risk includes del 17p, t(4;14),), t(14;16), t(14;20).
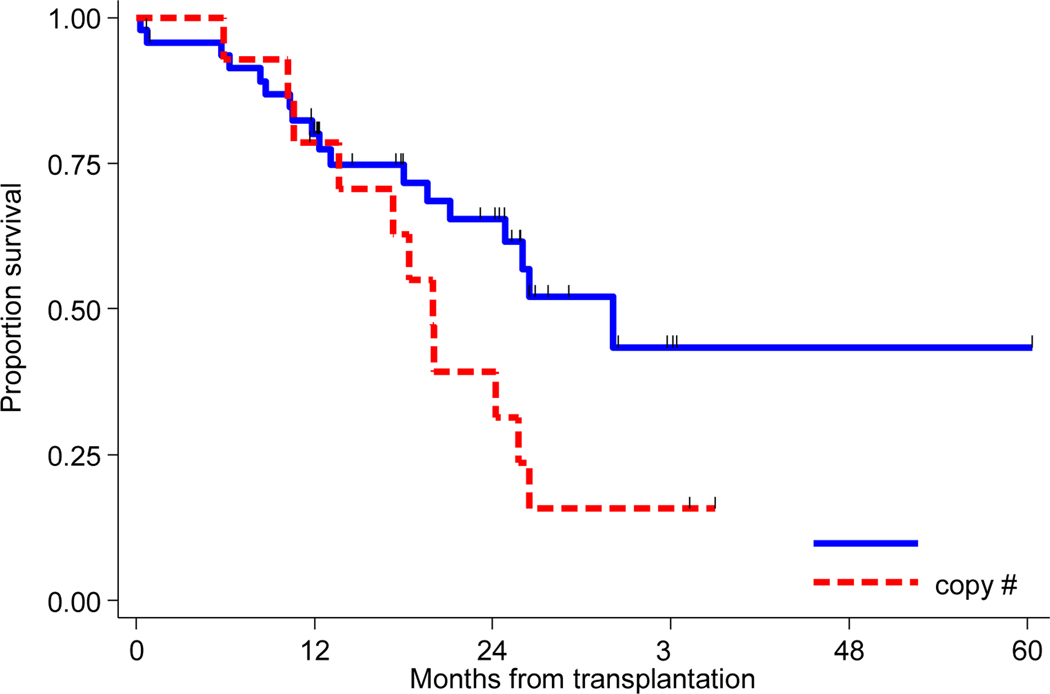
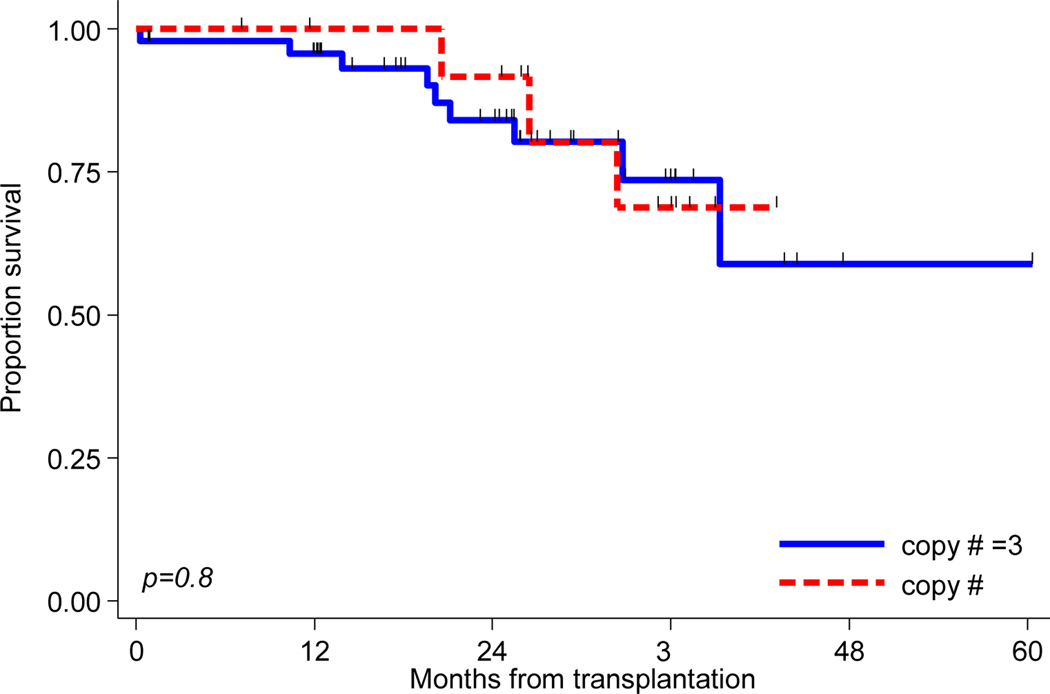
Impact of copy number variation on outcomes in patients with 1q+ was also analyszed. Information about copy number for 1q+ was available for 62 of the 96 1q+patients (65%). Forty-eight (50%) patients had 3 copies of 1q+ and 14 (15%) patients had ≥ 4 copies. The median PFS for patients with 3 copies of 1q+was 32.1 months compared to 20.0 months for patients who had ≥4 copies (Figure 2A). The association between PFS and copy number group approached significance (HR 0.49, CI 0.23 −1.05, P=0.06). Median OS had not reached for either group (Figure 2B), and no significant association between OS and copy number group was found (P=0.84).
Figure 2A:
Kaplan-Meier curves of progression free survival for 3 (N= 48) and ≥4 copies (N=14) of 1q+
Figure 2B:
Kaplan-Meier curves of overall survival for 3 (N= 48) and ≥4 copies (N=14) of 1q+
DISCUSSION
Abnormalities of chromosome 1 play an important role in the progression of MM. Here, we reported the clinical characteristics and outcomes of 1q+/1p− in newly diagnosed MM patients who received an IMiD or PI-based induction and upfront high dose therapy and auto-HCT. Most of these patients also received an IMiD or PI-based maintenance therapy. Presence of 1q+/1p− was associated with inferior PFS when compared to a propensity-score matched group of MM patients with diploid cytogenetics and normal FISH panel. However, the ORR, NRM, and OS were similar between the two groups. Our study also suggested an independent effect of 1q+/1p− on survival after auto-HCT. The statistical significance could not be reached perhaps due to the smaller sample size after propensity score matching of the 1q+/1p− alone (with no additional high-risk cytogenetics) and the controls.
Historically, the median PFS of newly diagnosed 1q+ MM patients has been reported to be less than two years after auto-HCT12,24. Kazmi et al. reported the median PFS of 10.8 months in high-risk MM patients from our center after auto-HCT 3. However, that study did not report the PFS for individual high-risk abnormalities and included patients who had their auto-HCT between 2005–2009, when triplet induction and post-transplant maintenance therapy was not the standard. Only 20% of the high-risk patients in that study received maintenance therapy and around one third of the patient received induction therapy with just one novel agent. Shah et.al recently reported the interim findings of the randomized-controlled Myeloma XI trial, which compared different novel induction, consolidation and maintenance regimens in newly diagnosed MM 25. They reported a median PFS of 19 months for 1q+ MM patients. However, not every patient in the study received auto-HCT, and some of the patients were randomized to a no-maintenance arm. Also, the PFS of patients who underwent auto-HCT and received maintenance therapy was not reported separately. The median PFS of our 1q+/1p− patients was 26.5 months, which is better than the historical controls12,24 and consistent with several recent studies where patients received induction and maintenance therapy with novel agents 13,26. Eighty-five percent of our patients received triplet-based induction (60% with PI and IMiD), all received auto-HCT and more than 70% received maintenance therapy. Additionally, 13% of our 1q+/1p− MM patients received Busulfan and Melphalan (Bu-Mel) as conditioning regimen for auto-HCT. Based on the available data from different studies, use of a triplet induction27,28, an effective conditioning29–31 and maintenance therapy32–34 may further overcome the negative impact of 1q+/1p− abnormalities.
Fonseca et al. reported that 1q+ by itself is not an independent prognostic marker and its effect on survival was dependent on the presence of additional high-risk cytogenetic abnormalities 14. Their study was also retrospective and included patients who received auto-HCT both upfront and at relapse. Several other prospective and retrospective studies 21,24,26 have shown the independent impact of 1q+ on survival after auto-HCT. Our study also suggested an independent effect of 1q+ on the outcome after auto-HCT (HR 1.9). The statistical significance could not be reached perhaps due to smaller sample size of patients with 1q+1/p- (N=58) after propensity matching. Our study was strengthened by the fact that all of our patients were uniformly treated, underwent upfront auto- HCT and we used a propensity score-matched control group which helped us in minimizing potential bias due to non-randomization.
Impact of copy number variation of 1q+ on survival has been reported in a few studies with contradictory results. An et al. reported comparable PFS and OS in MM patients with three or more than three copies of 1q+35. Hanamura et al. also showed that variations in copy number of 1q+ did not affect the 5 year event-free survival or OS 21. However, Neben et al. reported that both the PFS and OS were worse in patients with > 3 copies when compared to the group that had ≤ 3 copies of 1q+ 26. We did not see a difference in OS in patients with 3 versus ≥ 4 copies of 1q+, but there was a trend towards worse PFS in patients who had ≥ 4 copies of 1q+.
The University of Arkansas for Medical Science group, using microarray analysis on tumor cells of newly diagnosed MM identified a 70 gene signature which was associated with early progression and 30% of those genes were mapped to chromosome one 20. Most of the upregulated genes were mapped to chromosome 1q and the down-regulated genes were mapped to chromosome 1p. The three commonly deleted regions of 1p are 1p12 (FAM46C), 1p22.1 (RPL5) and 1p32.3 (CDKN2C). All of these are associated with inferior survival, however, the association with OS is most significant for 1p32.3 36. Median PFS of 26.5 months in our study was better than that reported by others for 1p- 17,18. This could be due to a small number of patients with 1p-, the use of novel induction regimens and post-transplantation maintenance therapy in our patients.
Our study has the usual limitations of a retrospective chart review, and the incidence of 1q+/1p− reported here is lower than what was reported in other studies. This could be due to the fact that we only included those MM patients who underwent upfront auto-HCT. Furthermore, in some patients, the high-risk myeloma FISH panel was not performed at the time of diagnosis, but after the initiation of induction treatment, which may have lowered the detection rate for 1q+/1p−. Additionally, not all bone marrow aspirate samples were enriched for CD138+ plasma cells, which could have also led to a lower detection rate for 1q+/1p−. We wanted to include the minimal residual disease information (MRD) for our patients, an important surrogate marker for survival after auto-HCT37. However, our patients were transplanted between 2006 and 2015 and the sensitivity of the assay used for the measurement of MRD was suboptimal when compared to current standards, therefore, it would not have added any useful information to this study.
In summary, chromosome 1 abnormalities (1q+/1p−) were associated with a worse outcome and should be considered a marker for high risk in MM. The outcomes of 1q+1/p- MM patients have improved with the use of more effective induction, conditioning, and maintenance therapy compared to historical controls, but they still need more effective therapeutic approaches to fully overcome the negative impact of 1q+1/p-. Future studies should also focus on the incorporation of newer agents, including monoclonal antibodies, chimeric antigen receptor T cells, and bispecific antibodies into the treatment of this high-risk group.
Highlights.
CKS1B gain(1q+) /CDKN2C deletion (1p-) have shorter PFS despite propensity score matching
PFS of 3 copies of 1q+ was longer than ≥4 copies and approached significance
1q+/1p− was suggestive of being an independent risk factor for survival
Acknowledgments
Funding Support /Financial disclosure: There are no financial disclosures
Footnotes
Conflict of Interest: There are no relevant conflict of interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Scott EC, Hari P, Sharma M, et al. Post-Transplant Outcomes in High-Risk Compared with Non-High-Risk Multiple Myeloma: A CIBMTR Analysis. Biol Blood Marrow Transplant. 2016;22(10):1893–1899. doi: 10.1016/j.bbmt.2016.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cornell RF, D’Souza A, Kassim AA, et al. Maintenance versus Induction Therapy Choice on Outcomes after Autologous Transplantation for Multiple Myeloma. Biol Blood Marrow Transplant. 2017;23(2):269–277. doi: 10.1016/j.bbmt.2016.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kazmi SM, Nusrat M, Gunaydin H, et al. Outcomes Among High-Risk and Standard-Risk Multiple Myeloma Patients Treated With High-Dose Chemotherapy and Autologous Hematopoietic Stem-Cell Transplantation. Clin Lymphoma Myeloma Leuk. 2015;15(11):687–693. doi: 10.1016/j.clml.2015.07.641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chng WJ, Dispenzieri A, Chim C-S, et al. IMWG consensus on risk stratification in multiple myeloma. Leukemia. 2014;28(2):269–277. doi: 10.1038/leu.2013.247 [DOI] [PubMed] [Google Scholar]
- 5.Scott EC, Hari P, Kumar S, et al. Staging Systems for Newly Diagnosed Myeloma Patients Undergoing Autologous Hematopoietic Cell Transplantation: The Revised International Staging System Shows the Most Differentiation between Groups. Biol Blood Marrow Transplant. 2018;24(12):2443–2449. doi: 10.1016/j.bbmt.2018.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gopalakrishnan S, D’Souza A, Scott E, et al. Revised International Staging System Is Predictive and Prognostic for Early Relapse (<24 months) after Autologous Transplantation for Newly Diagnosed Multiple Myeloma. Biol Blood Marrow Transplant. 2019;25(4):683–688. doi: 10.1016/j.bbmt.2018.12.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rajan AM, Rajkumar S V. Interpretation of cytogenetic results in multiple myeloma for clinical practice. Blood Cancer J. 2015;5:e365. doi: 10.1038/bcj.2015.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spruck C, Strohmaier H, Watson M, et al. A CDK-independent function of mammalian Cks1: targeting of SCF(Skp2) to the CDK inhibitor p27Kip1. Mol Cell. 2001;7(3):639–650. http://www.ncbi.nlm.nih.gov/pubmed/11463388. [DOI] [PubMed] [Google Scholar]
- 9.Ganoth D, Bornstein G, Ko TK, et al. The cell-cycle regulatory protein Cks1 is required for SCF(Skp2)-mediated ubiquitinylation of p27. Nat Cell Biol. 2001;3(3):321–324. doi: 10.1038/35060126 [DOI] [PubMed] [Google Scholar]
- 10.Shaughnessy J. Amplification and overexpression of CKS1B at chromosome band 1q21 is associated with reduced levels of p27Kip1 and an aggressive clinical course in multiple myeloma. Hematology. 2005;10 Suppl 1:117–126. doi: 10.1080/10245330512331390140 [DOI] [PubMed] [Google Scholar]
- 11.Bock F, Lu G, Srour SA, et al. Outcome of Patients with Multiple Myeloma and CKS1B Gene Amplification after Autologous Hematopoietic Stem Cell Transplantation. Biol Blood Marrow Transplant. 2016;22(12):2159–2164. doi: 10.1016/j.bbmt.2016.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang H, Qi X, Trieu Y, et al. Multiple myeloma patients with CKS1B gene amplification have a shorter progression-free survival post-autologous stem cell transplantation. Br J Haematol. 2006;135(4):486–491. doi: 10.1111/j.1365-2141.2006.06325.x [DOI] [PubMed] [Google Scholar]
- 13.Shah GL, Landau H, Londono D, et al. Gain of chromosome 1q portends worse prognosis in multiple myeloma despite novel agent-based induction regimens and autologous transplantation. Leuk Lymphoma. 2017;58(8):1823–1831. doi: 10.1080/10428194.2016.1260126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fonseca R, Van Wier SA, Chng WJ, et al. Prognostic value of chromosome 1q21 gain by fluorescent in situ hybridization and increase CKS1B expression in myeloma. Leukemia. 2006;20(11):2034–2040. doi: 10.1038/sj.leu.2404403 [DOI] [PubMed] [Google Scholar]
- 15.Leone PE, Walker BA, Jenner MW, et al. Deletions of CDKN2C in multiple myeloma: biological and clinical implications. Clin Cancer Res. 2008;14(19):6033–6041. doi: 10.1158/1078-0432.CCR-08-0347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qazilbash MH, Saliba RM, Ahmed B, et al. Deletion of the short arm of chromosome 1 (del 1p) is a strong predictor of poor outcome in myeloma patients undergoing an autotransplant. Biol Blood Marrow Transplant. 2007;13(9):1066–1072. doi: 10.1016/j.bbmt.2007.05.014 [DOI] [PubMed] [Google Scholar]
- 17.Boyd KD, Ross FM, Walker BA, et al. Mapping of chromosome 1p deletions in myeloma identifies FAM46C at 1p12 and CDKN2C at 1p32.3 as being genes in regions associated with adverse survival. Clin Cancer Res. 2011;17(24):7776–7784. doi: 10.1158/1078-0432.CCR-11-1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hebraud B, Leleu X, Lauwers-Cances V, et al. Deletion of the 1p32 region is a major independent prognostic factor in young patients with myeloma: the IFM experience on 1195 patients. Leukemia. 2014;28(3):675–679. doi: 10.1038/leu.2013.225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Avet-Loiseau H, Leleu X, Roussel M, et al. Bortezomib plus dexamethasone induction improves outcome of patients with t(4;14) myeloma but not outcome of patients with del(17p). J Clin Oncol. 2010;28(30):4630–4634. doi: 10.1200/JCO.2010.28.3945 [DOI] [PubMed] [Google Scholar]
- 20.Shaughnessy JD, Zhan F, Burington BE, et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood. 2007;109(6):2276–2284. doi: 10.1182/blood-2006-07-038430 [DOI] [PubMed] [Google Scholar]
- 21.Hanamura I, Stewart JP, Huang Y, et al. Frequent gain of chromosome band 1q21 in plasma-cell dyscrasias detected by fluorescence in situ hybridization: incidence increases from MGUS to relapsed myeloma and is related to prognosis and disease progression following tandem stem-cell transplantatio. Blood. 2006;108(5):1724–1732. doi: 10.1182/blood-2006-03-009910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar S, Paiva B, Anderson KC, et al. International Myeloma Working Group consensus criteria for response and minimal residual disease assessment in multiple myeloma. Lancet Oncol. 2016;17(8):e328–e346. doi: 10.1016/S1470-2045(16)30206-6 [DOI] [PubMed] [Google Scholar]
- 23.Garderet L, D’Souza A, Jacobs P, et al. Response Assessment in Myeloma: Practical Manual on Consistent Reporting in an Era of Dramatic Therapeutic Advances. Biol Blood Marrow Transplant. 2017;23(7):1193–1202. doi: 10.1016/j.bbmt.2017.03.009 [DOI] [PubMed] [Google Scholar]
- 24.Nemec P, Zemanova Z, Greslikova H, et al. Gain of 1q21 is an unfavorable genetic prognostic factor for multiple myeloma patients treated with high-dose chemotherapy. Biol Blood Marrow Transplant. 2010;16(4):548–554. doi: 10.1016/j.bbmt.2009.11.025 [DOI] [PubMed] [Google Scholar]
- 25.Shah V, Sherborne AL, Walker BA, et al. Prediction of outcome in newly diagnosed myeloma: a meta-analysis of the molecular profiles of 1905 trial patients. Leukemia. 2018;32(1):102–110. doi: 10.1038/leu.2017.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neben K, Lokhorst HM, Jauch A, et al. Administration of bortezomib before and after autologous stem cell transplantation improves outcome in multiple myeloma patients with deletion 17p. Blood. 2012;119(4):940–948. doi: 10.1182/blood-2011-09-379164 [DOI] [PubMed] [Google Scholar]
- 27.Durie BGM, Hoering A, Abidi MH, et al. Bortezomib with lenalidomide and dexamethasone versus lenalidomide and dexamethasone alone in patients with newly diagnosed myeloma without intent for immediate autologous stem-cell transplant (SWOG S0777): a randomised, open-label, phase 3 trial. Lancet (London, England). 2017;389(10068):519–527. doi: 10.1016/S0140-6736(16)31594-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosiñol L, Oriol A, Teruel AI, et al. Superiority of bortezomib, thalidomide, and dexamethasone (VTD) as induction pretransplantation therapy in multiple myeloma: a randomized phase 3 PETHEMA/GEM study. Blood. 2012;120(8):1589–1596. doi: 10.1182/blood-2012-02-408922 [DOI] [PubMed] [Google Scholar]
- 29.Bashir Q, Thall PF, Milton DR, et al. Conditioning with busulfan plus melphalan versus melphalan alone before autologous haemopoietic cell transplantation for multiple myeloma: an open-label, randomised, phase 3 trial. Lancet Haematol. March2019. doi: 10.1016/S2352-3026(19)30023-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jung S-H, Lee J-J, Kim JS, et al. Phase 2 Study of an Intravenous Busulfan and Melphalan Conditioning Regimen for Autologous Stem Cell Transplantation in Patients with Multiple Myeloma (KMM150). Biol Blood Marrow Transplant. 2018;24(5):923–929. doi: 10.1016/j.bbmt.2018.01.004 [DOI] [PubMed] [Google Scholar]
- 31.Hayman S, LaPlant B, Buadi F, et al. Phase 1/2 Trial of Carfilzomib and Melphalan Conditioning for Autologous Stem Cell Transplantation for Multiple Myeloma (CARAMEL). Biol Blood Marrow Transplant. 2019;25(3):S30. doi: 10.1016/j.bbmt.2018.12.102 [DOI] [Google Scholar]
- 32.Attal M, Lauwers-Cances V, Marit G, et al. Lenalidomide maintenance after stem-cell transplantation for multiple myeloma. N Engl J Med. 2012;366(19):1782–1791. doi: 10.1056/NEJMoa1114138 [DOI] [PubMed] [Google Scholar]
- 33.McCarthy PL, Owzar K, Hofmeister CC, et al. Lenalidomide after Stem-Cell Transplantation for Multiple Myeloma. N Engl J Med. 2012;366(19):1770–1781. doi: 10.1056/NEJMoa1114083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sivaraj D, Green MM, Li Z, et al. Outcomes of Maintenance Therapy with Bortezomib after Autologous Stem Cell Transplantation for Patients with Multiple Myeloma. Biol Blood Marrow Transplant. 2017;23(2):262–268. doi: 10.1016/j.bbmt.2016.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.An G, Xu Y, Shi L, et al. Chromosome 1q21 gains confer inferior outcomes in multiple myeloma treated with bortezomib but copy number variation and percentage of plasma cells involved have no additional prognostic value. Haematologica. 2014;99(2):353–359. doi: 10.3324/haematol.2013.088211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walker BA, Mavrommatis K, Wardell CP, et al. Identification of novel mutational drivers reveals oncogene dependencies in multiple myeloma. Blood. 2018;132(6):587–597. doi: 10.1182/blood-2018-03-840132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chakraborty R, Muchtar E, Kumar SK, et al. Impact of Post-Transplant Response and Minimal Residual Disease on Survival in Myeloma with High-Risk Cytogenetics. Biol Blood Marrow Transplant. 2017;23(4):598–605. doi: 10.1016/j.bbmt.2017.01.076 [DOI] [PubMed] [Google Scholar]