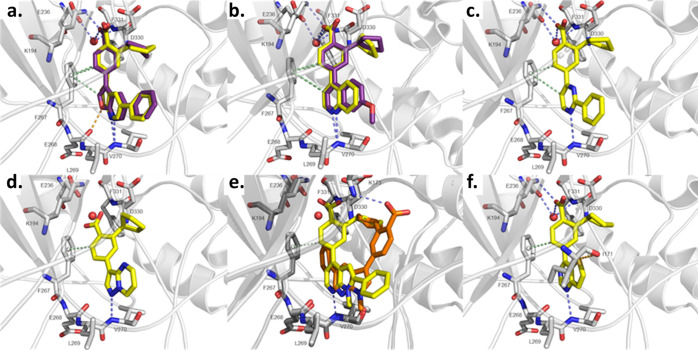
Figure 4.
X-ray structures and in silico docking. In silico docking was performed with Glide, and images were generated with PyMOL. The protein is colored in gray. Blue-dashed lines indicate H-bond interactions, while green-dashed lines display CH−π interactions and orange-dashed lines refer to sulfur−σ hole bonding. Docked ligands are shown as yellow sticks or orange sticks, co-crystallized ligands as purple sticks, and the water molecule as a red sphere. Oxygen and nitrogen atoms are colored in red and blue, respectively. (a) Predicted binding mode of 14g (yellow) using the protein structure from PDB-ID 5UY6 compared to the co-crystallized ligand 13g (purple). (b) Predicted binding modes of 46 (yellow) using the protein structure from PDB-ID 5UYJ and the co-crystallized ligand UNC10244803 (purple). (c) Predicted binding mode of 56 using the protein structure from PDB-ID 5UYJ. (d) Predicted binding mode of 22 using the protein structure from PDB-ID 6BKU. (e) Predicted binding modes of 10 (orange) and 29 (yellow) using the protein structure from PDB-ID 6BKU (light gray) and 5UYJ (dark gray), respectively. (f) Predicted binding mode of 38 using the protein structure from PDB-ID 5UYJ.