Abstract
4-Androstene-3,17-dione (4-AD), 1,4-androstadiene-3,17-dione (ADD) and 9α-hydroxyl-4-androstene-3,17-dione (9OH-AD), which are important starting compounds for the synthesis of steroidal medicines, can be biosynthetically transformed from phytosterols by Mycobacterium strains. Genomic and metabolic analyses have revealed that currently available 4-AD-producing strains maintain the ability to convert 4-AD to ADD and 9OH-AD via 3-ketosteroid-1,2-dehydrogenase (KstD) and 3-ketosteroid-9α-hydroxylase (Ksh), not only lowering the production yield of 4-AD but also hampering its purification refinement. Additionally, these 4-AD industrial strains are excellent model strains to construct ADD- and 9OH-AD-producing strains. We recently found that Mycobacterium neoaurum HGMS2, a 4-AD-producing strain, harbored fewer kstd and ksh genes through whole-genomic and enzymatic analyses, compared with other strains (Wang et al. in Microbial Cell Fact 19:187, 2020). In this study, we attempted to construct an efficient 4-AD-producing strain by knocking out the kstd and ksh genes from the M. neoaurum HGMS2 strain. Next, we used kstd- and ksh-default HGMS2 mutants as templates to construct ADD- and 9OH-AD-producing strains by knocking in active kstd and ksh genes, respectively. We found that after knocking out its endogenous kstd and ksh genes, one of these knockout mutants, HGMS2Δkstd211 + ΔkshB122, showed a 20% increase in the rate of phytosterol to 4-AD conversion, compared relative to the wild-type strain and an increase in 4-AD yield to 38.3 g/L in pilot-scale fermentation. Furthermore, we obtained the ADD- and 9OH-AD-producing strains, HGMS2kstd2 + Δkstd211+ΔkshB122 and HGMS2kshA51 + Δkstd211+ΔkshA226, by knocking in heterogenous active kstd and ksh genes to selected HGMS2 mutants, respectively. During pilot-scale fermentation, the conversion rates of the ADD- and 9OH-AD-producing mutants transforming phytosterol were 42.5 and 40.3%, respectively, and their yields reached 34.2 and 37.3 g/L, respectively. Overall, our study provides efficient strains for the production of 4-AD, ADD and 9OH-AD for the pharmaceutical industry and provides insights into the metabolic engineering of the HGMS2 strain to produce other important steroidal compounds.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12934-021-01653-9.
Keywords: 1,4-Androstadiene-3,17-dione (ADD); 21-Hydroxy-20-methylpregn-4-en-3-one (BA); 3-Ketosteroid-1,2-dehydrogenase (KstD); 3-Ketosteroid-9α-hydroxylase (Ksh); 4-Androstene-3,17-dione (4-AD); 9α-Hydroxyl-4-androstene-3,17-dione (9OH-AD); Mycobacterium sp. strain
Introduction
Steroids are widely used as antibacterial, anti-inflammatory, antiviral and anticancer therapeutics [1, 2]. The core structure of steroids consists of three six-member cyclohexane rings and one five-member cyclopentane ring. 4-Androstene-3,17-dione (4-AD), 1,4-androstadiene-3,17-dione (ADD) and 9α-hydroxyandrost-4-ene-3,17-dione (9OH-AD) are the most important starting materials for the synthesis of advanced steroid compounds. Accordingly, these three compounds are in high demanded by the pharmaceutical industry [3, 4]. The bioconversion of phytosterol to 4-AD, which is an important process in the pharmaceutical industry, uses bacteria that harbor partial steroid-degrading pathways [5, 6]. Although many microbes such as Pseudomonas sp. NCIB 10590 [7] and Nocardioides simplex VKM Ac-2033D [2, 8] contain steroid-degrading pathways, only strains from the Mycobacterium genus can efficiently accumulate 4-AD for potential industrial application [1, 3, 9, 10]. Mycobacterium neoaurum NRRL B-3805 was the first strain that was used to industrially transform phytosterol to 4-AD via its incomplete steroid-degrading pathways [11, 12]. Since then, other mycobacterial strains with the potential industrial application in the biotransformation of phytosterol to 4-AD, have been identified, including Mycobacterium sp. NwIB-01 [10]. Nevertheless, the accumulation of 4-AD in currently available industrial 4-AD-producing strains is often accompanied by the occurrence of ADD, 9OH-AD and other 4-AD derivatives generated during phytosterol transformation [1, 4]. The presence of these 4-AD derivates not only hampers the 4-AD purification and refining process but also significantly reduces 4-AD yield. However, no effective ADD- and 9OH-AD-producing strains are currently available for industrial-scale application.
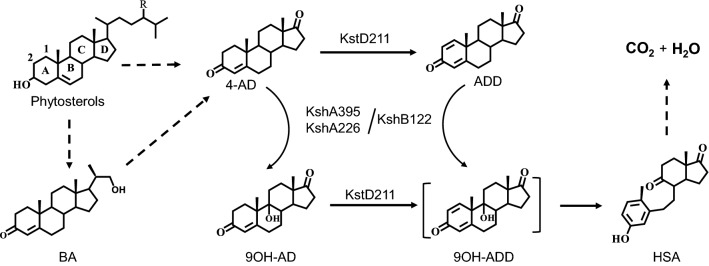
Biochemical and genomic investigations have revealed that all steroid-degrading bacteria share a core pathway via 9,10-secosteroid intermediates that participate in the breakdown of steroid ring structures [13, 14]. The conversion of phytosterol to 4-AD involves 11 enzymes in 14 consecutive enzymatic steps that must occurs before 4-AD can be completely degraded [15, 16]. Therefore, 4-AD can be considered a catabolic intermediate in the phytosterol-degrading pathway and a precursor of 9,10-secosteroid intermediate. Two enzymes, 3-ketosteroid-1,2-dehydrogenase (KstD) and 3-ketosteroid 9α-hydroxylase (Ksh), are responsible for the rupture of the 4-AD ring at the C9,10-position. This leads to the formation of 3-hydroxy-9,10-secoandrost-1,3,5(10)-triene-9,17-dione (HSA), which is readily catabolized to CO2 and H2O [17–19].
Reducing KstD and Ksh activities in 4-AD-producing strains has been shown to prevent the ring opening of steroidal intermediates and improve the accumulation of 4-AD, ADD and 9OH-AD during phytosterol transformation [20–32]. Traditional mutation approaches including ultra-violet light and chemical-mediated mutation have been employed to improve the productivity of 4-AD and other sterol compounds [33–39]. Owning to progress in the genome sequencing of sterol degradation strains, recent studies have focused on engineering highly efficient phytosterol-transforming strains using gene and metabolic engineering. Knockout of the casC gene from Rhodococcus jostii RHA1 resulted in the accumulation of a 5-carbon side chain cholate metabolite [40]. Additionally, depletion of the choD, hsdD and kstD genes in M. smegmatis vaccae enhanced the accumulation of 9OH-AD [33]. Knockout of the kstd gene from the M. neoaurum NwIB-01 strain increased AD and ADD production [10], while knockout of the kstd genes from M. neoaurum ATCC25795 harboring highly active Ksh enzymes led to the accumulation of 9OH-AD [23, 30]. However, to the best of our knowledge, these strains have not been reported for industrial application because of the existence of multiple and even unknown KstD and Ksh enzymes in these strains.
KstD is a flavin adenine dinucleotide (FAD)-dependent enzyme that introduces a double bond at the position between the C1 and C2 atoms of the A-ring of 3-ketosteroid substrates including 4-AD and 9OH-AD [41]. A bioinformatic study revealed that KstD enzymes exist in more than 500 different microbes, especially in bacteria belonging to the phylum Actinobacteria [41]. Moreover, a single species may contain multiple KstDs that are substrate specific. For instance, Rhizobium sp. strain [42], M. neoaurum DSM 1381 [43, 44] and Gordonia neofelifaecis NRRL B-59395 [45] carry at least three different KstD enzymes, each of which exhibits unique features in terms of its amino acid sequence, three-dimensional conformational structure and substrate specificity. Ksh is a two-component iron-sulfur monooxygenase that consists of the oxygenase KshA and the reductase KshB. The Ksh homologues are distributed over several phylogenetic groups [46]. For example, Mycobacterium sp. VKM Ac-1817D effectively transforms phytosterol into 9OH-AD using five KshAs and two KshBs [47]. Additionally, R. rhodochrous DSM43269 contains five KshA homologues, each of which exhibits distinct specificity for different substrates [48–50]. Many putative ksh genes have been identified in currently available 4-AD-producing strains [10, 51]. The balance between the accumulation of 4-AD and the inhibition of 4-AD degradation during phytosterol fermentation is managed by controlling the fermentation time. However, it is difficult to control this balance during prolonged fermentation processes, which often results in unstable conversion yield [16, 52]. This drawback becomes even worse when phytosterol fermentation is conducted in an aqueous medium [53, 54]. Accordingly, it is essential that an ideal 4-AD producing strain lacks all KstD and Ksh activities for industrial application.
We recently characterized a 4-AD producing strain, Mycobacterium neoaurum HGMS2, using genomic analysis and found it contained less kstd and ksh genes than other investigated strains. Specifically, the genome of the HGMS2 strain contains one kstd gene (kstd211), two kshA genes (kshA226 and kshA395) and one kshB gene (kshB122) [55]. The enzymes encoded by these genes, KstD211, KshA226, KshA395 and KshB122, are relevant to the generation of 4-ADD and 9OH-AD and the degradation of 4-AD during phytosterol fermentation (Fig. 1). In this study, we investigated the effects of these four genes on the accumulation of 4-AD during phytosterol transformation, using a homologous recombination approach, with the goal of providing efficient 4-AD-, ADD- and 9OH-AD-producing strains for industrial application. Knockout of the kstd211 and kshB122 genes from the HGMS2 strain abolished the accumulation of ADD and 9OH-AD and significantly improved the conversion yield of 4-AD compared with that of the wild-type strain. Furthermore, we knocked in heterogenous active kstd or ksh genes to selected HGMS2 mutants to generate ADD- and 9OH-AD-producing strains. As a result, our study work provided a high-efficiency 4-AD-producing bacterial strain and also provides ADD- and 9OH-AD-producing strains with potential application in the pharmaceutical industry.
Fig. 1.
KstD and Ksh enzymes are responsible for 4-AD accumulation and its degradation during phytosterol catabolism in M. neoaurum HGMS2. The steroidal core structure contains four rings, namely, rings A, B, C, and D, labeled on phytosterol. Successive steps involve the formation of 9α-hydroxy-1,4-androstadiene-3,17-dione (9OH-ADD), which is a transient intermediate and simultaneously converts to 3-hydroxy-9,10-secoandrost-1,3,5(10)-triene-9,17-dione (HSA). 4-AD: 4-androstene-3,17-dione; ADD: 1,4-androstadiene-3,17-dione; 9OH-AD: 9α-hydroxy-4-androstene-3,17-dione and BA: 21-hydroxy-20-methylpregn-4-en-3-one
Materials and methods
Materials
A DNA gel extraction kit was purchased from Omega Biotek (Hubei, China). Other molecular biology reagents were of the highest grade and were obtained from Thermo Scientific (Shanghai, China). 4-Androstene-3,17-dione (4-AD), 1,4-androstene-3,17-dione (ADD), 9α-hydroxyl-4-androstene-3,17-dione (9OH-AD), 21-hydroxy-20-methylpregn-4-en-3-one (BA) and phytosterol were obtained from Hubei Goto Pharmaceutical Co. (Xiangyang, China). In this study, the purity of phytosterol obtained from commercial source was 95% on average and typically, composed of β-sitosterol (47.5%, MW = 414.71 Da), campesterol (26.4%, MW = 402.7 Da), stigmasterol (17.7%, MW = 412.69 Da) and brassicasterol (3.6%, MW = 398.66 Da) and so that an average of molecular weight of 410.40 Da. Phenazine methosulfate (PMS, 99%) and nitro blue tetrazolium with purities of 99% were purchased from Sigma-Aldrich (Shanghai, China). Restriction enzymes, dNTPs, and Taq polymerase were purchased from TaKaRa Co. (Dalian, China).
Bacterial strains, plasmids, DNA extraction and purification
Mycobacterium neoaurum HGMS2, a non-pathogenic industrial Mycobacterium, was maintained in our laboratory and deposited at China Center for Type Culture Collection (CCTCC No: M2012522) [55] and its genome sequence was deposited in GenBank (CP031414.1). The plasmids p2NIL (Cat. 20188) [56] and pT18mobsacB (Cat. 72648) were purchased from AddGene (Watertown, MA, USA).
The genomic DNAs of Mycobacterium sp. HGMS2 and its mutants were extracted using a Bacterial Genomic DNA Extraction Kit from Tiangen (Beijing, China). Plasmids were purified using a Plasmid Purification Kit from Tiangen (Beijing, China). PCR fragments were purified using an agarose gel purification kit (Qiagen, USA).
Construction of homogenous recombinant vectors and knockout and knockin experiment
The pT18mobsacB was used as a template to amplify the sucrose lethality gene SacB that was originally from Bacillus subtilis 168. SacB and its promotor and terminator were amplified by PCR using two primers (sacB-F and sacB-R) that introduced EcoRI sites at both ends. The nucleotide sequences of sacB-F and sacB-R were 5′-CCGGAATTCCACATATACCTGCCGTTC-3′ and 5′-CCGGAATTCTTATTTGTTAACTGTTAATTGT-3′, respectively. The PCR fragment and the p2NIL plasmid were individually digested with the EcoRI restriction enzyme. The SacB fragment and the restriction plasmid were recovered by agarose gel purification. Digested SacB DNA fragment was subcloned into the digested p2NIL plasmid by T4 DNA ligase, and the resultant plasmid was named p2NILSac.
To knock out the kstd and ksh genes from the HGMS2 genome, each of these genes was replaced through homologous recombination with a specific DNA fragment that contained the upstream sequence and the downstream sequence outside of the targeted gene. Each upstream sequence and each downstream sequence were approximately 1 kbp in length. The upstream sequence and the downstream sequence of each target gene were amplified from the HGMS2 genome (Additional file 1: Table S1) using two pairs of primers (Additional file 1: Table S2). The amplified upstream sequence and downstream sequence for each target gene were digested with two pairs of restriction enzymes, BamHI/XbaI and XbaI/HindIII, respectively. Digested fragments were ligated into the p2NILSac vector that was predigested with BamH I/HindIII. The ligated plasmids were transformed into E. coli DH5a competent cells for amplification. Constructed plasmids were confirmed by DNA sequencing.
The same strategy was used to knock in active kstd and ksh genes. The kstd2 gene from Mycobacterium neoaurum DSM 1381 (Additional file 1: Figure S1) [44] was amplified by PCR using the primers KstD-OF and KstD-OR (Additional file 1: Table S3). The amplified kstd2 fragment and the plasmid p2NIL-SacB with kstd upstream and downstream homology arms were treated with XbaI and digested products were purified with PCR clean-up kits. The digested kstd2 gene fragment and plasmid were ligated using T4 DNA ligase to obtain the KstD2 knockin vector. The ligation mixture was transformed into E. coli DH5a for amplification and DNA sequencing. The kshA knockin vector was constructed in the same manner as the above except for its template. A kshA gene was adopted from kshA51 (Additional file 1: Figure S2) [57] and chemically synthesized. The synthetic kshA51 gene was ligated with the plasmid p2NIL-SacB with kshA226 upstream and downstream homology arms to obtain the kshA51 knockin vector.
Screening of recombinant mutants
DNA electroporation was performed with a Gene Pulser Xcell System (BioRad, Hercules, CA, USA) according to a protocol described by Goude et al. [58]. The competent cells were prepared by the following procedure. The bacteria were cultured in 50 mL of LB medium containing 0.05% Tween-80 until the OD600nm value reached 0.4. The cells were collected by centrifugation and washed three times with 10% glycerol containing 0.05% Tween-80. Cell pellets were then resuspended in 100 µL of 10% glycerol solution and used as competent cells. Then, 5 ng of recombinant plasmid was added to 100 µL of competent cells, and the mixture was transferred to a 0.2 cm precooled electroporation cuvette. A single pulse of 2.5 kV, 25 µF and a resistance of 1000 Ω with a constant wave pulse was usually used. The time constant was set for 23 ms. The bacterial suspension was diluted to 1 mL with LB medium and incubated for 3 h at 30 °C. The culture was then spread on an LB agar plate containing 50 mg/ml kanamycin (LBK) and allowed to grow at 30 °C for 3 days. Colonies were screened by PCR for positive recombination.
A few colonies growing on LBK plates were picked, and each colony was inoculated into 5 mL of LBT medium (0.05 Tween-80 in LB) containing 50 mg/mL of kanamycin and cultured at 30 °C and 200 rpm for 2 days. The cell culture was diluted in 103- and 105-fold with LBT medium. Then, 100 µL of diluted cell suspension was spread on LBK agar plates containing 10% sucrose (LBKS), and 100 µL of diluted cell suspension was also spread on LBK agar plates as a control. Both types of plates were incubated at 30 °C for 3 days. When the number of colonies on the LBKS plates was less than that on the LBK plates, colonies were picked from the LBKS plates for PCR verification. Positive colonies were transferred to 5 mL of LBT medium and cultured at 30 °C and 200 rpm for 2 days. Their genomic DNAs were extracted for PCR verification. The verified mutant strains were stored at − 80 °C.
Small-scale fermentation
The wild-type and mutant Mycobacterium strains were initially cultured in 5 mL of LB medium at 30 °C for two or three days till its OD600nm value reached 13–15. The culture is used as seeds and was inoculated into a 100 mL of fermentation medium that contains yeast extract (8 g/L), glycerol (6 g/L), (NH4)2HPO4 (0.6 g/L), NaNO3 (2 g/L), phytosterol (10 g/L), F-35 (20 g/L), β-cyclodextrin (5 g/L), and lectin (3 g/L), and shaken at 30 °C and 200 rpm. Then, 1 ml of fermentation broth was collected every 24 h for 7 days for the extraction of metabolites.
The fermentation broth was thoroughly mixed with ethyl acetate at a ratio of 1:1. The mixture was centrifuged at 8000×g for 5 min and the supernatant was collected. An aliquot of the supernatant was directly used for thin layer chromatographic (TLC) assay. The supernatant was collected and dried by heating using a hair drier. The dried sample was dissolved in 40% methanol solution for high-performance liquid chromatographic (HPLC) assays.
Molecular cloning, expression and purification of KstD2 and KshA51 enzymes
Preparation of KstD211 and KshA395 were described previously by Wang et al. [55]. The synthetic KstD2 and KshA51 genes were subcloned through BamH I and EcoR I into a modified pET28b vector that its thrombin cleavage site was replaced with a Tobacco Etch Virus (TEV) protease cleavage site. Each plasmid was individually transformed into E. coli BL21(DE3) cells for protein expression. Protein expression and purification of KstD2 and KshA51 enzymes were performed according to previously-reported procedure [55]. Purified KstD2 and KshA51 enzymes were assayed with SDS-PAGE as a single band and used for activity assay.
3-Ketosteroid-1,2-dehydrogenase activity assay
The activity of 3-ketosteroid-1,2-dehydrogenase (KstD) was measured spectrophotometrically at 600 nm for 10 min at 30 °C using 5-Methylphenazinium methosulfate (PMS) and 2,6-Dichlorophenolindphenol (DCPIP) as artificial electron acceptors. The reaction was conducted in 500 µL of mixture solution containing 50 mM Tris-HCl buffer (pH 7.0), 1.5 mM PMS, 40 µM DCPIP and 250 mM AD in 2% methanol. The reaction was initiated by adding 20 µg of purified KstD211 or KstD2. A reaction lacking AD was monitored as a control. Activity is expressed as U/mg of protein after subtracting control data. One unit (U) of enzyme activity is defined as the amount of enzyme required for reduction of 1 µmol of DCPIP per minute (ε600 nm = 18.7 × 103 L/mol·cm).
3-Ketosteroid-9a-hydroxylase activity assay
The assay of 3-ketosteroid-9a-hydroxylase (Ksh) activity was performed by a method modified from a procedure published by Petrusma et al. [50]. Briefly, the reaction mixture contained 500 µL of air-saturated 50 mM Tris-HCl buffer (pH 7.0), 100 µM NADH and 20 µg of enzyme. The reaction was initiated by the addition of 250 µM 4-AD into the mixture. The substrate solution was prepared by dissolving 4-AD powder in methanol. All assays were performed at 25 °C. A kinetic program on a NanoDrop C2000 spectrophotometer was used to continuously record NADH oxidation at 340 nm (ε = 6.22 L/mol cm).
The enzymatic conversion of 4-AD to 9OH-AD was also evaluated by monitoring substrate and product concentrations using an HPLC system equipped with a Waters 2945 UV detector at 215 nm and a 150 × 4.6 mm C18 reverse-phase 5 μm ODS-analytical column. The mobile phase contained 40% methanol and 60% water and the flow rate was set at 1 ml/min. In this work, the elution times for 4-AD and 9OH-AD were 10.6 and 4.4 min, respectively. The concentrations of 4-AD and 9OH-AD were calculated from their respective peak areas using standard curves of 4-AD and 9OH-AD, respectively. The R2 value for the standard curve was 0.99.
TLC and HPLC analyses of fermentation metabolites
TLC assays were performed on 0.25-mm-thick silica gel that impregnated with fluorescent dye (Haiyang Chemical Co., Qingdao, China). Sample extracts were spotted in 5 µL aliquots onto silica TLC plates. The TLC plates were developed with a solvent system containing petroleum ether and ethyl acetate at a ratio of 6:4 (v/v) as the mobile phase. The products of the enzymatic reaction on TLC plates were visualized under ultraviolet (UV) light. If needed, the TLC plates were stained with 20% sulfuric acid at 100 °C for 10 min to identify compounds that were invisible under UV light.
HPLC analysis was carried out on a C18 reverse-phase Sunfire column (5 μm, 4.6 × 150 mm, Waters, USA) at 30 °C with a Waters HPLC system equipped with a UV detector. Extracted metabolites dissolved in ethyl acetate were transferred to a clean glass vial and dried in vacuo. Dried powder was redissolved in methanol (approximately 1 mg/mL), and the solution was then filtered through a 0.22 μm microporous membrane. HPLC was performed using a mixture of methanol and water at a ratio of 60:40 (v/v) as the mobile phase at a flow rate of 1 mL/min. Analytes were detected with UV light at 214 nm. The identities of HPLC peaks were confirmed by comparison with the standards of 4-AD, ADD, and 9OH-AD. Each peak area was integrated with software provided by Waters and used to evaluate the compound concentration.
The conversion rate (Conv) of phytosterol to 4-AD, ADD and 9OH-AD was estimated using the following equation:
where Mst and Mp are the weights of steroid and phytosterol, respectively. MWst and MWp are the molecular weights of steroid and phytosterol, respectively. In this work, the average molecular weight of phytosterol is 410.40 Da (see above).
Mass spectrometric analysis of fermentation metabolites
LC/MS mass spectra of HPLC fractions were obtained under isocratic or gradient elution conditions, with acetonitrile:water (1:1) as the mobile phase. Small percentages of formic acid were added to the water mobile phase in some experiments to check the effects of the additives on the sensitivities and the mass spectra of the steroid compounds. After choosing the best conditions, i.e., acetonitrile:water = 60:40 with 0.2% formic acid, the detection limit was assessed for each fraction using selected ion monitoring of the most abundant ions in the spectra.
Pilot-scale fermentation
Pilot-scale fermentation was conducted in a 15 L fermenter. The fermentation medium consisted of yeast extract (8 g/L), glycerol (6 g/L), (NH4)2HPO4 (0.6 g/L), NaNO3 (2 g/L), phytosterol (80 g/L), F-35 (20 g/L), β-cyclodextrin (20 g/L) and lectin (3 g/L). Phytosterol was mixed with glycerol and β-cyclodextrin, and thoroughly emulsified before the mixture was transferred to the fermenter. All of the other materials were added into the fermenter, and water was added until the total volume of the medium reached 10 L. The medium was autoclaved in situ at 121 °C for 30 min, followed by cooling down to 30 °C with stirring at 500 rpm. The fermentation medium was inoculated with 1% (v/v) of the seed that was cultured for 2 days. The fermentation was processed at 30 °C with stirring at 500 rpm and the dissolved O2 concentration (50–60%) and the pH value (pH 7.5) of the fermentation broth were constantly monitored in-line and automatically adjusted if necessary. Samples were collected every 6 h to monitor the concentration of 4-AD, ADD and 9OH-AD.
Results
Characterization of HGMS2 fermentation extracts
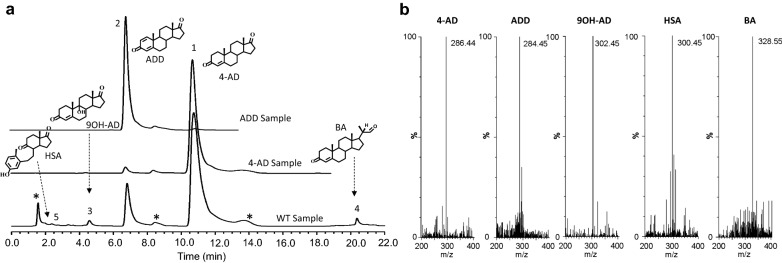
The wild-type HGSM2 strain has been used in the pharmaceutical industry to produce 4-AD with a reasonable conversion rate and yield using phytosterol as a substate in an 80 m3 fermenter. A typical fermentation process was carried out for 7–10 days, during which time the process was monitored by TLC and HPLC assays until the substrates were completely converted. Figure 2a shows the HPLC profile of an extract from the HGMS2 fermentation broth after seven days of phytosterol fermentation. As shown in Fig. 2a, multiple products were accumulated during fermentation. Two major products were 4-AD and ADD, which were readily identified by comparison with the HPLC profiles of the standard 4-AD and ADD samples. Other minor peaks were identified as 9OH-AD, HSA and BA. All five products were further confirmed by mass spectrometry (Fig. 2b). Based on HPLC profiling, the contents of the five products were 78.21%, 10.33%, 1.43%, 3.85 and 0.54% for 4-AD, ADD, 9OH-AD BA and HSA, respectively, whereas the remaining contents were unknown metabolites unrelated to phytosterol catabolism.
Fig. 2.
Characterization of major catabolites of the extracts from large-scale HGMS2 fermentation using phytosterol as sole carbon source for 144 h. a HPLC profiling of the extracts in comparison with those of standard 4-AD and ADD samples. (1) 4-AD; (2) ADD; (3) 9OH-AD; (4) BA; asterisk: unknown HGMS2 metabolites. b Mass spectra of the five major HPLC peaks
Construction of KstD-knockout and Ksh-knockout mutants
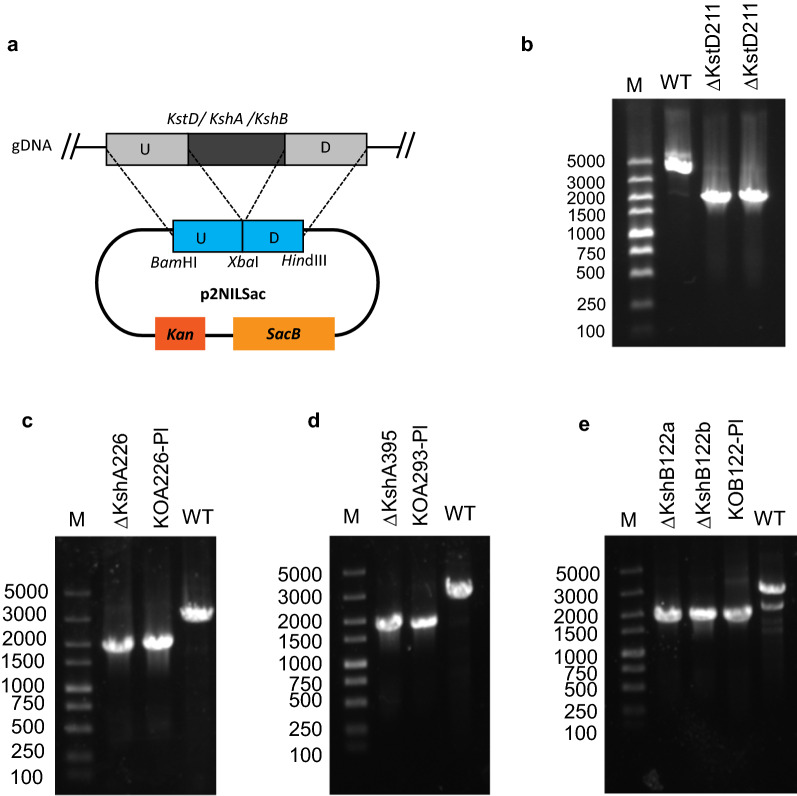
As indicated above, active KstD and Ksh enzymes diminished the accumulation of 4-AD via the formation of 9,10-secosteroid during phytosterol fermentation by the HGMS2 strain. Thus, we attempted to knock out these genes from the genome of the HGMS2 strain by homologous recombination approaches. To construct a homologous recombination vector with an appropriate antibiotic marker, we examined whether the HGMS2 strain had the ability to resist common antibiotics that can be deactivated by bacterial proteins. Although many mycobacterial species are resistant to a variety of antibiotics [59, 60], as shown in Additional file 1: Table S4, we found that the HGMS2 strain was not resistant to any tested antibiotics including kanamycin (Kan), tetracycline (Tc), streptomycin (Stm), spectinomycin (Spm), ampicillin (Amp) and zeocin (Zn). Thus, we chose Kan as the first selection marker and the sucrose lethality gene (SacB) [61] as the secondary selection marker to construct gene knockout/knockin vectors. Using the p2NIL vector that contains a Kan gene as a template, we inserted a synthetic SacB gene into the p2NIL vector behind the Kan gene. The resultant vector was named p2NILSacB (Fig. 3a).
Fig. 3.
Knockout of the kstd and ksh genes from the HGMS2 strain. a A homologous recombination vector was constructed based on p2NIL plasmid. U: the upstream sequence; D: the downstream sequence. b PCR confirmation of putative kstd-knockout colonies, in comparison with that for the WT strain. M: DNA marker; WT: PCR products amplified from the HGMS2; ΔKstD211: two putative kstd-default mutants. c–e PCR confirmation of putative ksh-knockout colonies, in comparison with that for the WT strain. M: DNA marker; ΔKshA226: a putative HGMS2ΔkstD211 + ΔkshA226 mutant; KOA226-Pl: the plasmid p2NIL-Sac-ΔKshA226; WT: the HGMS2 strain; ΔKshA395: a putative HGMS2Δkstd211 + ΔkshA395 mutant; KOA395-Pl: the plasmid p2NIL-Sac-ΔKshA395; ΔKshB122a and ΔKshB122: two putative HGMS2Δkstd211 + ΔkshB122 mutants; KOB122-Pl: the plasmid p2NIL-Sac-ΔKshB122
To use the pNILSac vector to knock out one kstd211 and three ksh genes from the HGMS2 strain, each DNA fragment containing around 1000 bp from both sides of a target gene was amplified with PCR and inserted into the BamH I and Hind III sites in the p2NILSac vector. Four recombinant vectors were constructed to delete the kstd211, kshA226, kshA395 and kshB122 genes from the HGMS2 genome and then confirmed by PCR (Additional file 1: Figure S3) and DNA sequencing. The four vectors were named p2NIL-SacB-ΔKstD211, p2NIL-SacB-ΔKshA226, p2NIL-SacB-ΔKshA395 and p2NILSacB-ΔKshB122 (Table 1).
Table 1.
Lists of homologous recombinant knockout and knockin vectors
| Mutation | Vector | Targeted gene | Starting strain |
|---|---|---|---|
| KO | p2NIL-SacB-ΔKstD211 | kstd211 | WT |
| KO | p2NIL-Sacb-ΔKshA226 | kshA226 | HGMS2Δkstd211 |
| KO | p2NIL-SacB-ΔKshA395 | kshA395 | HGMS2Δkstd211 |
| KO | p2NIL-SacB-ΔKshB122 | kshB122 | HGMS2Δkstd211 |
| KI | p2NIL-SacB-KstD2 | kstD211 |
HGMS2Δkstd211 ΔkshB122 |
| KI | p2NIL-SacB-KshA51 | kshA226 | HGMS2Δkstd211 |
Because ADD was a major by-product produced by active KstD211 enzyme during phytosterol fermentation with the wild-type HGMS2 strain, we knocked out the kstd211 gene from the HGMS2 genome in the first stage and then sequentially knocked out ksh genes from the kstd-knockout mutant. The knockout of the kstd211 gene was done by electroporation of the p2NIL-SacB-ΔKstD plasmid into fresh HGMS2 competent cells, followed by a two-step selection with kan- and sucrose-based methods (see “Materials and methods”). Positive recombinant colonies were identified with colony PCR. After the kstd211 gene was deleted from the HGMS2 genome, the size of the PCR product became much shorter (1600 bp) than that of the wild-type strain (3600 bp). As shown in Fig. 3b, two positive colonies were identified, and DNA sequencing results confirmed that the kstd211 gene was knocked out from both clones. We arbitrarily selected one of these two colonies for further investigation and named it HGMS2Δkstd211.
As mentioned above, the HGMS2 strain contains two pairs of KshA/KshB enzyme components sharing one single KshB enzyme. To explore the importance of individual Ksh enzymes in the formation of 9OH-AD, we knocked out each ksh gene from the HGMS2Δkstd211 mutant using the same knockout strategy that was used togenerate the HGMS2Δkstd211 mutant. As shown in Fig. 3c and e, positive recombinant colonies were identified by colony PCR. DNA sequencing also confirmed that these ksh genes were removed from the three colonies. The resultant mutants were named HGMS2Δkstd211+ΔkshA226, HGMS2Δkstd211+ΔkshA395 and HGMS2Δkstd211+ΔkshB122.
Effect of KstD211 and Ksh enzymes on HGMS2 phytosterol transformation
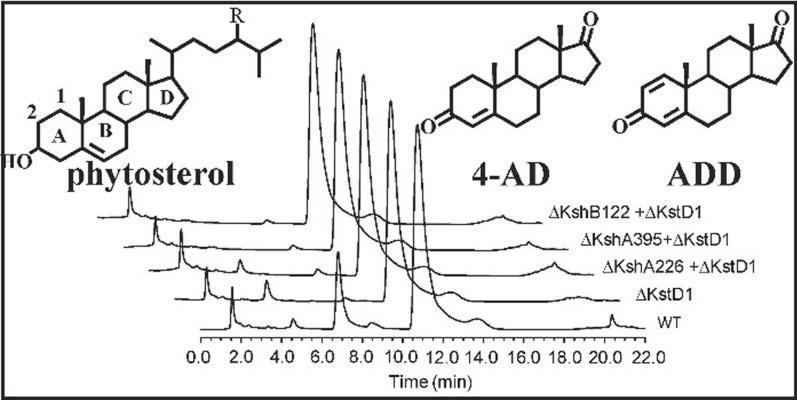
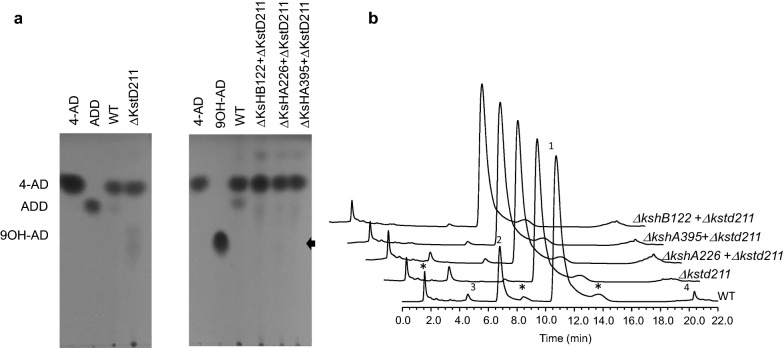
To evaluate the effects of kstd211 and ksh knockouts on phytosterol transformation in the HGMS2 strain, small-scale phytosterol fermentation was carried out for the HGMS2 strain and its knockout mutants. First, we tested the HGMS2Δkstd211 mutant. After phytosterol fermentation for 7 days, the fermentation broth was extracted with a solvent and evaluated byTLC assay. As shown in Fig. 4a, the HGMS2Δkstd211 mutant abolished ADD production compared with that of the WT strain. However, 9OH-AD was detected on the TLC plate. Notably, knockout of the kstd211 gene slightly increased the conversion of 4-AD to 9OH-AD (Fig. 4a). Furthermore, the extract was diluted to the same concentration as that of the WT sample based on the TLC density and subjected to HPLC assay. As shown in Fig. 4b, knockout of the kstd211 gene caused the HGMS2Δkstd211 strain to be almost free of ADD (peak 2 in Fig. 4b), and we once again observed that the yield of 9OH-AD increased (peak 3 in Fig. 4b). It was reasonable that the accumulation of 9OH-AD occurred when the KstD211-involved formation of 9,10-secosteroid was blocked.
Fig. 4.
Characterization of kstd-knockout and ksh-knockout mutants for phytosterol fermentation in small-scale fermentation. a TLC assay of the catabolites by kstd-knockout and ksh-knockout mutants during phytosterol fermentation, compared with those of the WT strain and the standard samples of 4-AD, ADD and 9OH-AD. ΔKstD1, ΔKshB122, ΔKshA226 and ΔKshA395 refer to the HGMS2Δkstd211, HGMS2Δkstd211 + ΔkshB122, HGMS2Δkstd211 + ΔkshA226 and HGMS2Δkstd211 + ΔkshA395 mutants, respectively. Weak 9OH-AD spots for the samples from the HGMS2Δkstd211, and HGMS2Δkstd211 + ΔkshA226 mutants are marked by arrows. b HPLC profile of the extracts from the phytosterol fermentation by three double mutants for 144 h compared with those of the WT strain and the HGMS2Δkstd211 mutant. (1) 4-AD; (2) ADD; (3) 9OH-AD and (4) BA. Asterisk: unknown HGMS2 metabolites when the strain was cultured without phytosterol
We further tested the effects of the three ksh knockout mutants on phytosterol transformation on a small scale. As shown in Fig. 4a, knockout of the three ksh genes from the HGMS2 strain exerted a distinct effect on the accumulation of 9OH-AD. A weak spot was found for the sample from the HGMS2Δkstd211+ΔkshA226 mutant, while no detecTable 9OH-AD spots were visualized for the other two mutants. This observation was consistent with our previous finding that KshA226 was inactive for 4-AD production, in contrast to KshA395 and KshB122 [55]. As shown in Fig. 4b, HPLC profiling of these extracts clearly indicated that the production of 9OH-AD was completely blocked during phytosterol transformation in the HGMS2Δkstd211+ΔkshA395 and HGMS2Δkstd211+ΔkshB122 mutants. Notably, the production of ADD was also blocked in these two mutant strains. No detecTable 9OH-AD was observed at the end of 7 days of fermentation and 4-AD significantly accumulated in the HGMS2Δkstd1+ΔkshB122 mutant with a conversion rate of 47.2% and a yield of 5.62 g/L when starting from 10 g/L of phytosterol (Table 2). Because the deletion of kshB122 could simultaneously disable the functions of the Ksh226 and KshA395 enzymes, we selected the HGMS2Δkstd1+ΔkshB122 mutant for pilot scale fermentation.
Table 2.
Comparison of conversion rates and yields of 4-AD, ADD and 9OH-AD from phytosterol by a variety of Mycobacterium strains
| Strain | Phytosterol concentration (g/L) |
Yield | Conversion rate | References | ||
|---|---|---|---|---|---|---|
| 4-AD (g/L) | ADD (g/L) | 9OH-AD (g/L) | ||||
| 4-AD-producing strains | ||||||
| Mycobacterium sp. ATCC 25795 | 15 | 6.85 | 5.57 | N/A | 45.67% (4-AD); 37.13% (ADD) | [64] |
| Mycobacterium sp. B-3683 | 5 | 2.35 | N/A | N/A | 47% | [65] |
| Mycobacterium sp. B-3805 | 20 | 7.4 | N/A | N/A | 37% | [51] |
| Mycobacterium smegmatis MC2 155 | 10 | 8.8-9.0 | 1.0-1.1 | N/A | N/A | [26] |
| Mycobacterium sp. HGMS2 | 10 | 5.62 | 1.30 | 0.30 | 47.2% | This work |
| Mycobacterium sp. HGMS2 | 80 | 31.4 | 6.26 | 0.88 | 40.6% | This work |
| Mycobacterium sp. HGMS2Δkstd211 + ΔkshB122 | 10 | 6.22 | NO | NO | 51.6% | This work |
| Mycobacterium sp. HGMS2Δkstd211 + ΔkshB122 | 80 | 38.3 | NO | NO | 48.7% | This work |
| ADD-producing strains | ||||||
| Mycobacterium neoaurum NwIB-04 | 15 | 0.096 | 4.94 | N/A | 32.93% | [32] |
| Mycobacterium neoaurum NwIB-01 | 0.4 | N/A | 0.174 | N/A | 43.5% | [31] |
| Mycobacterium sp. HGMS2kstd2 + Δkstd211+ΔkshB122 | 10 | NO | 4.12 | NO | 44.2% | This work |
| Mycobacterium sp. HGMS2kstd2 + Δkstd211+ΔkshB122 | 80 | NO | 34.2 | NO | 42.5% | This work |
| 9OH-AD-producing strains | ||||||
| Mycobacterium sp. MS136 | 13 | N/A | N/A | 6.0 | 40% | [21] |
| Mycobacterium neoaurum 25795 mutant | 20 | N/A | N/A | 10.27 | 51.2% | [63] |
| Mycobacterium neoaurum ATCC 25795 mutant | 15 | N/A | N/A | 6.20 | 41.33% | [30] |
| Mycobacterium sp. HGMS2kshA51 + Δkstd211+ΔkshA226 | 10 | NO | NO | 4.2 | 44.2% | This work |
| Mycobacterium sp. HGMS2kshA51 + Δkstd211+ΔkshA226 | 80 | NO | NO | 37.2 | 40.3% | This work |
N/A data not available, SD significant detectable, NO not observable
Pilot-scale phytosterol fermentation with the HGMS2Δkstd211+ΔkshB122 mutant
Pilot-scale phytosterol fermentations using both the wild-type HGMS2 strain and the HGMS2Δkstd211+ΔkshB122 mutant were conducted in a 15 L fermenter supplied with 10 L fermentation medium (see “Materials and methods”).
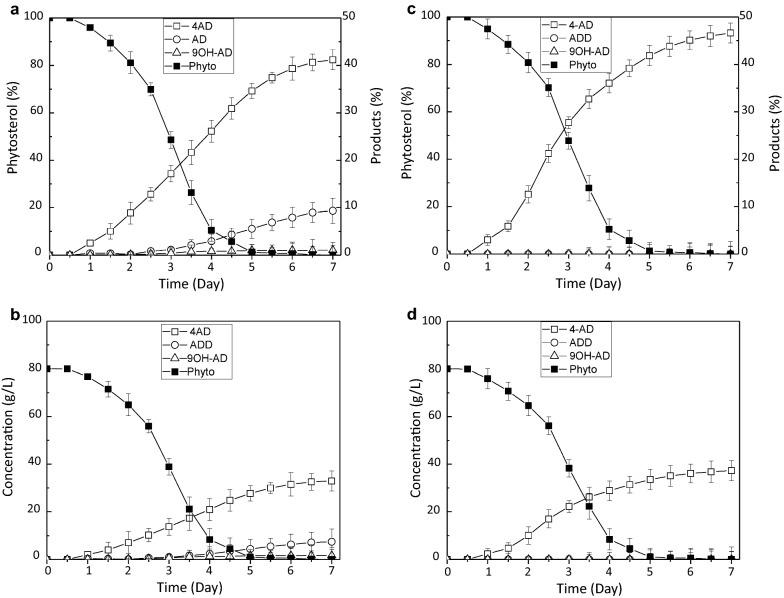
As shown in Fig. 5a, the rate of phytosterol conversion to 4-AD increased dramatically within the first 5 days and reached 40.6% after 7 days of fermentation with 80 g/L of phytosterol, while the phytosterol content in the medium decreased. On average, the final yield of 4-AD was 31.4 ± 4.3 g/L (Fig. 5b; Table 2). Accompanying the production of 4-AD was the accumulation of ADD and 9OH-AD also observed With yields of 6.26 g/L and 0.88 g/L on average (Table 2), respectively, after 7 days of fermentation (Fig. 5b), Resulting in total contents of 9.8 and 1.8% on average, respectively. (Fig. 5a).
Fig. 5.
Time course of phytosterol transformation to 4-AD, ADD and 9OH-AD in pilot-scale fermentation. a, b The conversion rate and yield of phytosterol by the HGMS2 strain, compared with the wild-type strain.; c, b The conversion rate and yield of phytosterol by the HGMS2ΔKstD211 + ΔkshB122 mutant
As expected, the HGMS2Δkstd211 + ΔkshB122 mutant exhibited an enhanced conversion rate of phytosterol to 4-AD. As shown in Fig. 5c, the rate of phytosterol conversion to 4-AD increased to 48.7% after 7days of fermentation. The final 4-AD yield in the fermentation broth was estimated to be 38.3 ± 4.7 g/L on average (Fig. 5d; Table 2), after transformation from 80 g/L of phytosterol. Notably, ADD and 9OH-AD almost completely disappeared. When compared with the wild-type HGMS2 strain, the mutant strain enhanced the conversion rate and 4-AD yield by 1.2-fold and 1.2-fold, respectively. Moreover, the HGMS2Δkstd211 + ΔkshB122 mutant completely catabolized the substrate with the same efficiency as the wild-type strain after 7 days of fermentation (Fig. 5).
Construction of ADD-producing and 9OH-AD-producing mutants
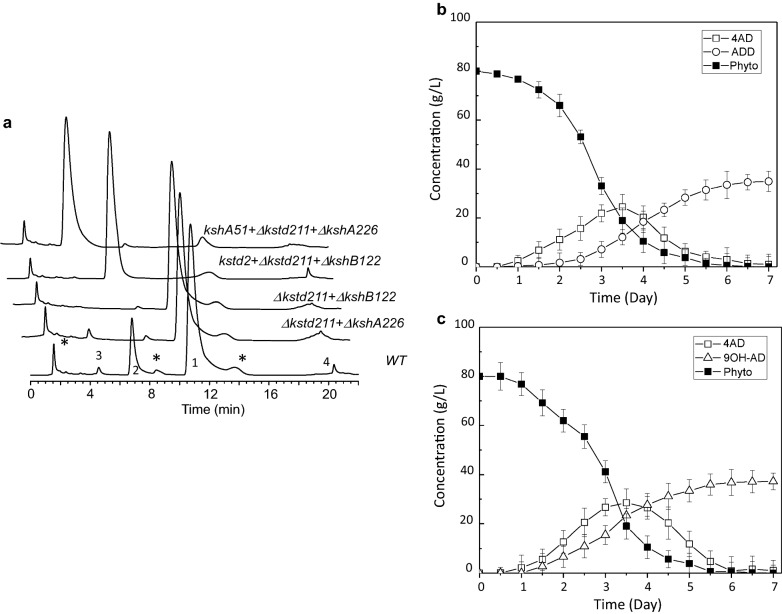
Encouraged by the performance of the HGMS2Δkstd211 + ΔkshB122 mutant, we attempted to use kstd- and ksh-default mutants as cell models to construct an ADD-producing strain and a 9OH-AD-producing strain. The KstD2 from Mycobacterium neoaurum DSM 1381 is homologous to KstD211 and other KstD enzymes (Additional file 1: Figure S4). The activity of the KstD2 enzyme is much higher than that of the original KstD enzyme in the HGMS2 strain (i.e., KstD211 that converts 4-AD to ADD;Table 3). Therefore, we substituted the kstd211 gene in HGMS2Δkstd1 + ΔkshB122 with the kstd2 gene to generate an ADD-producing mutant strain. This knockin mutant, which was confirmed by PCR (Additional file 1: Figure S5a) and DNA sequencing, was named HGMS2kstd2 + Δkstd211+ΔkshB122 (Table 3). During tested in a small-scale fermentation system using phytosterol as substrate, the HGMS2kstd2 + Δkstd211+ΔkshB122 mutant efficiently accumulated ADD within 7 days with a conversion rate of 44.2% and a yield of 4.12 g/L starting from 10 g/L of phytosterol (Table 2). Upon HPLC analysis (Fig. 6a), no detecTable 4-AD and 9OH-AD peaks were observed and all 4-AD generated in the HGMS2Δkstd211 + ΔkshB122 mutant was eventually transformed to ADD in the HGMS2kstd2 + Δkstd211+ΔkshB122 mutant. During pilot-scale fermentation, the conversion rate of phytosterol to ADD was 42.5% on average after 7 days of fermentation with 80 g/L of phytosterol. The final ADD yield in the fermentation broth was estimated to be 34.2 ± 5.3 g/L on average (Fig. 6b; Table 2). As shown in Fig. 6b, this ADD-producing mutant completely catabolized the substrate after 5.5 days during pilot-scale fermentation. In this process, 4-AD was an intermediate, and its accumulation started to increase within the first 3.5 days, after which it decreased until complete transformation to ADD had occurred.
Table 3.
Construction of ADD- and 9OH-AD-producing mutants through knocking in highly-active kstd and ksh genes, respectively
| Product | Starting strain | Original gene | Mutant strain | Knockin gene | ||
|---|---|---|---|---|---|---|
| Enzyme | In vitro Activity (units/mg) | Enzyme | In vitro activity (units/mg) | |||
| ADD | HGMS2Δkstkd211 + ΔkshB122 | KstD211 | 125.7 ± 23.4 | HGMS2Δkstd211 + ΔkshB122+kstD2 | KstD2 | 9238.7 ± 68 |
| 9OH-AD | HGMS2Δkstd211 + Δksh226 | KshA395 | 56.4 ± 7.9 | HGMS2Δkstd211 + ΔkshA226+kshA51 | KshA51 | 532 ± 42 |
Fig. 6.
Characterization of ADD- and 9OH-AD-producing mutants during phytosterol fermentation. a HPLC profile of the extracts from the phytosterol fermentation by ADD- and 9OH-AD-producing mutants for 144 h compared with those of HGMS2Δkstd211 + ΔkshA226 and the HGMS2Δkstd211 + ΔkshB122 mutants. (1) 4-AD; (2) ADD; (3) 9OH-AD and (4) BA. *: unknown HGMS2 metabolites when the strain was cultured without phytosterol. b, c Time course of ADD and 9OH-AD yields by the HGMS2kstd2 + Δkstd1+ΔkshA226 and the HGMS2kshA51 + Δkstd211+ΔkshB122 mutants, respectively
KshA51 is a chimeric KshA enzyme that is generated by KshA1 and KshA5 from R. rhodochrous DSM 43269 [57] and is homologous to KshA395 and other KshA enzymes (Additional file 1: Figure S6). KshA51 is much more active than KshA395 for 4-AD (Table 3). To construct a 9OH-AD-producing strain, we replaced the latent kshA226 gene in the HGMS2Δkstd211 + ΔkshA226 mutant with a synthetic KshA51 gene that was knocked into the locus of kshA226 in the HGMS2Δkstd211 + ΔkshA226 mutant. The resultant mutant was confirmed by PCR (Additional file 1: Figure S5b) and DNA sequencing and then named HGMS2kshA51 + Δkstd211+ΔkshA226 (Table 3). We examined the HGMS2kshA51 + Δkstd211+ΔkshA226 mutant for phytosterol transformation during small-scale fermentation for 7 days. The catabolites from the fermentation broth were extracted and evaluated by HPLC. As shown in Fig. 6a, no detecTable 4-AD or ADD was observed at the end of 7 days of fermentation, and 9OH-AD significantly accumulated in this mutant with a conversion rate of 44.2% and a yield of 4.2 g/L when starting from 10 g/L of phytosterol (Table 2). During a pilot-scale fermentation the conversion rate of phytosterol to 9OH-AD was 40.3% on average after 7 days of fermentation (Fig. 6c). As shown in Fig. 6c; Table 2, this 9OH-AD-producing mutant completely catabolized the phytosterol substrate (80 g/L) after 6 days, with an estimated final 9OH-AD yield in the fermentation broth of 37.2 ± 5.3 g/L on average. During phytosterol transformation, 4-AD was an intermediate and its 4-AD accumulation started to increase within the first 3 days, after which it decreased until early completely transformation to 9OH-AD had occurred.
Discussion
Previously, many Mycobacteria and their kstd-knockout and/or ksh-knockout mutants were investigated for the accumulation of 4-AD, ADD and/or 9OH-AD. Many of these strains exhibited reasonable rates for the transformation of phytosterol with the potential for industrial application (Table 2). However, two major issues remained. Specifically, these Mycobacterium strains either continued to produce multiple products that hampered product purification or were investigated at lower concentrations of phytosterol. This indicated that it was likely that the template strains used in these pioneer works contained residual kstd and ksh genes. Conversely, Mycobacterium neoaurum HGMS2 contained fewer kstd and ksh genes than other bacterial strains [26, 31, 43, 52, 62]. In this study, we first generated a few M. neoaurum HGMS2 mutants by knocking out the kstd and ksh genes. When compared with the wild-type strain, the mutants exhibited improved performance in the conversion of phytosterol to 4-AD. After the kstd211 gene was knocked out from the HGMS2 strain, the mutant HGMS2Δkstd211 strain lost its ability to convert 4-AD to ADD; thus, ADD disappeared from the phytosterol fermentation broth. Furthermore, three ksh genes were individually knocked out from the HGMS2Δkstd211 mutant. The three double mutants that were exhibited different abilities to convert 4-AD to 9OH-AD. Specifically, the HGMS2Δkstd211 + ΔkshA395 and HGMS2Δkstd211 + ΔkshB122 mutants completely blocked the occurrence of 9OH-AD, while the HGMS2Δkstd211 + ΔkshA226 mutant still converted 4-AD to 9OH-AD. Therefore, knocking out the kstd gene first and then the ksh genes benefited the accumulation of 4-AD during phytosterol conversion. Tested in pilot-scale fermentation with a high concentration of phytosterol revealed that the HGMS2Δkstd211 + ΔkshB122 mutant increased the conversion rate by 1.2-fold to 48.7% on average. resulting in the 4-AD yield transformed by this double mutant increasing to 38.3 ± 8.7 g/L. In our pilot-scale fermentation, the conversion rate and 4-AD yield of our new 4-AD-producing strain only reached 75% of their theoretical values, i.e., 65% and 52.0 g/L, respectively. As shown in Figs. 2a and 6a, it was likely that the HGMS2 and its mutants still transformed phytosterol to BA and other intermediates. Further gene engineering should improve the 4-AD production yield. In our pilot-scale study, we also observed that the viscosity of the fermentation broth increased significantly as the starting concentration of phytosterol increased from 10 to 80 g/L. Nevertheless, we did not observe any significant inhibitory growth by substrate or product (data not shown). Thus, we expect that it should be possible to increase the starting concentration of phytosterol in aqueous fermentation medium, when exploring new formulae for fermentation media.
Furthermore, we constructed ADD- and 9OH-AD-producing strains by knocking the genes of active heterogenous KstD and KshA enzymes into the M. neoaurum HGMS2 mutants, respectively. The resultant mutant HGMS2kstd2 + Δkstd211+ΔkshB122 could efficiently and completely convert phytosterol to ADD with an average conversion rate of 42.5% and an average yield of 34.2 ± 5.3 g/L from 80 g/L of phytosterol. The HGMS2kshA51 + Δkstd211+ΔkshA226 mutants also efficiently converted phytosterol into 9OH-AD with an average conversion rate of 40.3% and an average yield of 37.2 ± 5.2 g/L from 80 g/L of phytosterol. These results were comparable to those obtained with other mycobacterial strains [21, 30, 31, 63]. One of the significant advantages of our mutants was that they generated less impurities. Because these two mutants were made of kstd- and ksh-knockout strains, further knock outs of other impurity-related genes should improve the ADD and 9OH-AD production yield.
In conclusion, our work provides efficient mutants for the production of 4-AD, ADD and 9OH-AD for industrial application. The gene implementation of M. sp. HGMS2 in this work has demonstrated that the HGMS2 strain is an excellent model for engineering microbial cell factories to produce important steroidal compounds. The production of another important pharmaceutical compound, BA, via similar gene engineering methods is currently under investigation in our laboratory.
Supplementary Information
Additional file 1. Supplementary informations include homologous recombinant sequences for knocking out targeted genes in the HGMS2 mutants, primers used for the construction of knockout and knockin vectors, antibiotic resistances of the HGMS2 strain, DNA sequence of KstD2 gene, DNA sequence of KshA51 gene, amino acid alignment of KstD211 with other KstD enzymes, amino acid alignment of KshA51 with the Ksh enzymes from the HGMS2 strain, and experimental data for PCR screening of KO and KI mutants.
Acknowledgements
We thank Mr. Fangxin Lu for his technical assistance and Rita Su from the University of Toronto for language editing.
Abbreviations
- 4-AD
4-Androstene-3,17-dione
- 9OH-AD
9α-Hydroxyl-4-androstene-3,17-dione
- ADD
1,4-Androstene-3,17-dione
- BA
21-Hydroxy-20-methylpregn-4-en-3-one
- DCPIP
2,6-Dichlorophenolindophenol
- HPLC
High-performance liquid chromatography
- HSA
3-Hydroxy-9,10-secoandrost-1,3,5(10)-triene-9,17-dione
- KstD
3-Ketosteroid-1,2-dehydrogenase
- Ksh
3-Ketosteroid-9α-hydroxylase
- TLC
Thin layer chromatography
Authors’ contributions
ZDS, YJH and YQH conceived and designed research. XL, TC, FP, SKS, JPY, DNS and XYC conducted experiments. XL, FP, SKS and JPY analyzed data. ZDS, XL and YQH wrote the manuscript. All authors read and approved the manuscript.
Funding
The Hubei Provincial Ministry of Technology (ZDS, No: 2016ACA128), Natural Science Foundation of Hubei Province (Grant Number 2019CFB713) and National “111” Center for Cellular Regulation and Molecular Pharmaceutics (grant XBTK-2018001).
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xin Li, Tian Chen and Fei Peng contributed equally to this work
Contributor Information
Yijun He, Email: heyijun@gotochem.com.
Zhengding Su, Email: zhengdingsu@hbut.edu.cn.
References
- 1.Donova MV, Egorova OV. Microbial steroid transformations: current state and prospects. Appl Microbiol Biotechnol. 2012;94:1423–47. doi: 10.1007/s00253-012-4078-0. [DOI] [PubMed] [Google Scholar]
- 2.Shtratnikova VY, Schelkunov MI, Pekov YA, Fokina VV, Logacheva MD, Sokolov SL, Bragin EY, Ashapkin VV, Donova MV. Complete genome sequence of steroid-transforming nocardioides simplex VKM Ac-2033D. Genome Announc. 2015;3:e01406–14. doi: 10.1128/genomeA.01406-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernández-Cabezón L, Galán B, García JL. New insights on steroid biotechnology. Front Microbiol. 2018;9:15. doi: 10.3389/fmicb.2018.00958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donova MV. Steroid bioconversions. Methods Mol Biol. 2017;1645:1–13. doi: 10.1007/978-1-4939-7183-1_1. [DOI] [PubMed] [Google Scholar]
- 5.Fernandes P, Cruz A, Angelova B, Pinheiro HM, Cabral JMS. Microbial conversion of steroid compounds: recent developments. Enzyme Microb Technol. 2003;32:688–705. doi: 10.1016/S0141-0229(03)00029-2. [DOI] [Google Scholar]
- 6.Donova MV. Transformation of steroids by actinobacteria: a review. Appl Biochem Microbiol. 2007;43:1–14. doi: 10.1134/S0003683807010012. [DOI] [PubMed] [Google Scholar]
- 7.Owen RW, Mason AN, Bilton RF. The degradation of beta-sitosterol by Pseudomonas sp. NCIB 10590 under aerobic conditions. J Steroid Biochem. 1985;23:327–32. doi: 10.1016/0022-4731(85)90412-1. [DOI] [PubMed] [Google Scholar]
- 8.Shtratnikova VY, Schelkunov MI, Fokina VV, Pekov YA, Ivashina T, Donova MV. Genome-wide bioinformatics analysis of steroid metabolism-associated genes in Nocardioides simplex VKM Ac-2033D. Curr Genet. 2016;62:643–56. doi: 10.1007/s00294-016-0568-4. [DOI] [PubMed] [Google Scholar]
- 9.Wei JH, Yin X, Welander PV. Sterol synthesis in diverse bacteria. Front Microbiol. 2016;7:19. doi: 10.3389/fmicb.2016.00990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei W, Fan S, Wang F, Wei D. A new steroid-transforming strain of Mycobacterium neoaurum and cloning of 3-ketosteroid 9alpha-hydroxylase in NwIB-01. Appl Biochem Biotechnol. 2010;162:1446–56. doi: 10.1007/s12010-010-8919-y. [DOI] [PubMed] [Google Scholar]
- 11.Marsheck WJ, Kraychy S, Muir RD. Microbial degradation of sterols. Appl Microbiol. 1972;23:72–7. doi: 10.1128/am.23.1.72-77.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goren T, Harni M, Rimonk S, Aharonowitz Y. 1-Ene-steroid reductase of Mycobacterium SP. NRRL B-3805. J Steroid Biochem. 1983;19:1789–97. doi: 10.1016/0022-4731(83)90361-8. [DOI] [PubMed] [Google Scholar]
- 13.Holert J, Cardenas E, Bergstrand LH, Zaikova E, Hahn AS, Hallam SJ, Mohn WW. Metagenomes reveal global distribution of bacterial steroid catabolism in natural, engineered, and host environments. mBio. 2018;9:e02345-02317. doi: 10.1128/mBio.02345-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bergstrand LH, Cardenas E, Holert J, Van Hamme JD, Mohn WW. Delineation of steroid-degrading microorganisms through comparative genomic analysis. MBio. 2016;7:e00166. doi: 10.1128/mBio.00166-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujimoto Y, Chen CS, Szeleczky Z, Ditullio D, Sih CJ. Microbial degradation of the phytosterol side chain. I. Enzymic conversion of 3-oxo-24-ethylcholest-4-en-26-oic acid into 3-oxochol-4-en-24-oic acid and androst-4-ene-3,17-dione. J Am Chem Soc. 1982;104:4718–4720. doi: 10.1021/ja00381a055. [DOI] [Google Scholar]
- 16.Malaviya A, Gomes J. Androstenedione production by biotransformation of phytosterols. Bioresour Technol. 2008;99:6725–37. doi: 10.1016/j.biortech.2008.01.039. [DOI] [PubMed] [Google Scholar]
- 17.Kreit J. Microbial catabolism of sterols: focus on the enzymes that transform the sterol 3beta-hydroxy-5-en into 3-keto-4-en. FEMS Microbiol Lett. 2017;364:fnx007. doi: 10.1093/femsle/fnx007. [DOI] [PubMed] [Google Scholar]
- 18.Chiang Y-R, Wei ST-S, Wang P-H, Wu P-H, Yu C-P. Microbial degradation of steroid sex hormones: implications for environmental and ecological studies. Microb Biotechnol. 2020;13:926–49. doi: 10.1111/1751-7915.13504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kreit J. Aerobic catabolism of sterols by microorganisms: key enzymes that open the 3-ketosteroid nucleus. FEMS Microbiol Lett. 2019;366:fnz173. doi: 10.1093/femsle/fnz173. [DOI] [PubMed] [Google Scholar]
- 20.Li H, Wang X, Zhou L, Ma Y, Yuan W, Zhang X, Shi J, Xu Z. Enhancing expression of 3-ketosteroid-9α-hydroxylase oxygenase, an enzyme with broad substrate range and high hydroxylation ability, in Mycobacterium sp. LY-1. Appl Biochem Biotechnol. 2019;187:1238–54. doi: 10.1007/s12010-018-2876-2. [DOI] [PubMed] [Google Scholar]
- 21.He K, Sun H, Song H. Engineering phytosterol transport system in Mycobacterium sp. strain MS136 enhances production of 9α-hydroxy-4-androstene-3,17-dione. Biotech Lett. 2018;40:673–8. doi: 10.1007/s10529-018-2520-9. [DOI] [PubMed] [Google Scholar]
- 22.Shao M, Sha Z, Zhang X, Rao Z, Xu M, Yang T, Xu Z, Yang S. Efficient androst-1,4-diene-3,17-dione production by co-expressing 3-ketosteroid-∆1-dehydrogenase and catalase in Bacillus subtilis. J Appl Microbiol. 2017;122:119–28. doi: 10.1111/jam.13336. [DOI] [PubMed] [Google Scholar]
- 23.Liu HH, Xu LQ, Yao K, Xiong LB, Tao XY, Liu M, Wang Q, Wei DZ. Engineered 3-ketosteroid 9alpha-hydroxylases in emopenMycobacterium neoaurumemclose: an efficient platform for production of steroid drugs. Appl Environ Microbiol. 2018;84:e02777-17. doi: 10.1128/AEM.02777-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan S-F, Alper HS. Metabolic engineering of microbial cell factories for production of nutraceuticals. Microb Cell Fact. 2019;18:46. doi: 10.1186/s12934-019-1096-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.García-Fernández J, Martínez I, Fernández-Cabezón L, Felpeto-Santero C, García J-L, Galán B. Bioconversion of phytosterols into androstadienedione by Mycobacterium smegmatis CECT 8331. Methods Mol Biol (Clifton NJ) 2017;1645:211–25. doi: 10.1007/978-1-4939-7183-1_15. [DOI] [PubMed] [Google Scholar]
- 26.Galán B, Uhía I, García-Fernández E, Martínez I, Bahíllo E, de la Fuente JL, Barredo JL, Fernández-Cabezón L, García JL. Mycobacterium smegmatis is a suitable cell factory for the production of steroidic synthons. Microb Biotechnol. 2017;10:138–50. doi: 10.1111/1751-7915.12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Feng J, Zhang D, Wu Q, Zhu D, Ma Y. Characterization of new recombinant 3-ketosteroid-∆1-dehydrogenases for the biotransformation of steroids. Appl Microbiol Biotechnol. 2017;101:6049–60. doi: 10.1007/s00253-017-8378-2. [DOI] [PubMed] [Google Scholar]
- 28.Shao M, Zhang X, Rao Z, Xu M, Yang T, Li H, Xu Z, Yang S. A mutant form of 3-ketosteroid-Delta(1)-dehydrogenase gives altered androst-1,4-diene-3, 17-dione/androst-4-ene-3,17-dione molar ratios in steroid biotransformations by Mycobacterium neoaurum ST-095. J Ind Microbiol Biotechnol. 2016;43:691–701. doi: 10.1007/s10295-016-1743-9. [DOI] [PubMed] [Google Scholar]
- 29.Xie R, Shen Y, Qin N, Wang Y, Su L, Wang M. Genetic differences in ksdD influence on the ADD/AD ratio of Mycobacterium neoaurum. J Ind Microbiol Biotechnol. 2015;42:507–13. doi: 10.1007/s10295-014-1577-2. [DOI] [PubMed] [Google Scholar]
- 30.Yao K, Xu L-Q, Wang F-Q, Wei D-Z. Characterization and engineering of 3-ketosteroid-△1-dehydrogenase and 3-ketosteroid-9α-hydroxylase in Mycobacterium neoaurum ATCC 25795 to produce 9α-hydroxy-4-androstene-3,17-dione through the catabolism of sterols. Metab Eng. 2014;24:181–91. doi: 10.1016/j.ymben.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 31.Wei W, Fan S-Y, Wang F-Q, Wei D-Z. Accumulation of androstadiene-dione by overexpression of heterologous 3-ketosteroid ∆1-dehydrogenase in Mycobacterium neoaurum NwIB-01. World J Microbiol Biotechnol. 2014;30:1947–54. doi: 10.1007/s11274-014-1614-3. [DOI] [PubMed] [Google Scholar]
- 32.Wei W, Wang F-Q, Fan S-Y, Wei D-Z. Inactivation and augmentation of the primary 3-ketosteroid-{delta}1- dehydrogenase in Mycobacterium neoaurum NwIB-01: biotransformation of soybean phytosterols to 4-androstene- 3,17-dione or 1,4-androstadiene-3,17-dione. Appl Environ Microbiol. 2010;76:578–4582. doi: 10.1128/AEM.02088-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seidel L, Horhold C. Selection and characterization of new microorganisms for the manufacture of 9-OH-AD from sterols. J Basic Microbiol. 1992;32:49–55. doi: 10.1002/jobm.3620320115. [DOI] [PubMed] [Google Scholar]
- 34.Rodina NV, Molchanova MA, Voishvillo NE, Andryushina VA, Stytsenko TS. Conversion of phytosterols into androstenedione by Mycobacterium neoaurum. Appl Biochem Microbiol. 2008;44:48–54. doi: 10.1134/S0003683808010080. [DOI] [PubMed] [Google Scholar]
- 35.Su L, Shen Y, Xia M, Shang Z, Xu S, An X, Wang M. Overexpression of cytochrome p450 125 in Mycobacterium: a rational strategy in the promotion of phytosterol biotransformation. J Ind Microbiol Biotechnol. 2018;45:857–67. doi: 10.1007/s10295-018-2063-z. [DOI] [PubMed] [Google Scholar]
- 36.Su L, Shen Y, Zhang W, Gao T, Shang Z, Wang M. Cofactor engineering to regulate NAD(+)/NADH ratio with its application to phytosterols biotransformation. Microb Cell Fact. 2017;16:182. doi: 10.1186/s12934-017-0796-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Geize R, Hessels GI, van Gerwen R, van der Meijden P, Dijkhuizen L. Unmarked gene deletion mutagenesis of kstD, encoding 3-ketosteroid ∆1-dehydrogenase, in Rhodococcus erythropolis SQ1 using sacB as counter-selectable marker. FEMS Microbiol Lett. 2001;205:197–202. doi: 10.1111/j.1574-6968.2001.tb10947.x. [DOI] [PubMed] [Google Scholar]
- 38.van der Geize R, Hessels GI, van Gerwen R, Vrijbloed JW, van der Meijden P, Dijkhuizen L. Targeted disruption of the kstD gene encoding a 3-ketosteroid delta 1-dehydrogenase isoenzyme of Rhodococcus erythropolis strain SQ1. Appl Environ Microbiol. 2000;66:2029–36. doi: 10.1128/AEM.66.5.2029-2036.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petrusma M, van der Geize R, Dijkhuizen L. 3-Ketosteroid 9alpha-hydroxylase enzymes: rieske non-heme monooxygenases essential for bacterial steroid degradation. Antonie Van Leeuwenhoek. 2014;106:157–72. doi: 10.1007/s10482-014-0188-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruprecht A, Maddox J, Stirling AJ, Visaggio N, Seah SY. Characterization of novel acyl coenzyme A dehydrogenases involved in bacterial steroid degradation. J Bacteriol. 2015;197:1360–7. doi: 10.1128/JB.02420-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rohman A, Dijkstra BW. The role and mechanism of microbial 3-ketosteroid ∆1-dehydrogenases in steroid breakdown. J Steroid Biochem Mol Biol. 2019;191:105366. doi: 10.1016/j.jsbmb.2019.04.015. [DOI] [PubMed] [Google Scholar]
- 42.Guevara G, Heras LFdl, Perera J, Llorens JMN. Functional characterization of 3-ketosteroid 9α-hydroxylases in Rhodococcus ruber strain chol-4. J Steroid Biochem Mol Biol. 2017;172:176–87. doi: 10.1016/j.jsbmb.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 43.Zhang R, Liu X, Wang Y, Han Y, Sun J, Shi J, Zhang B. Identification, function, and application of 3-ketosteroid ∆1-dehydrogenase isozymes in Mycobacterium neoaurum DSM 1381 for the production of steroidic synthons. Microb Cell Fact. 2018;17:77. doi: 10.1186/s12934-018-0916-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang R, Xu X, Cao H, Yuan C, Yuminaga Y, Zhao S, Shi J, Zhang B. Purification, characterization, and application of a high activity 3-ketosteroid-Delta(1)-dehydrogenase from Mycobacterium neoaurum DSM 1381. Appl Microbiol Biotechnol. 2019;103:6605–16. doi: 10.1007/s00253-019-09988-5. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Q, Ren Y, He J, Cheng S, Yuan J, Ge F, Li W, Zhang Y, Xie G. Multiplicity of 3-ketosteroid ∆1-dehydrogenase enzymes in Gordonia neofelifaecis NRRL B-59395 with preferences for different steroids. Ann Microbiol. 2015;65:1961–71. doi: 10.1007/s13213-015-1034-0. [DOI] [Google Scholar]
- 46.Szaleniec M, Wojtkiewicz AM, Bernhardt R, Borowski T, Donova M. Bacterial steroid hydroxylases: enzyme classes, their functions and comparison of their catalytic mechanisms. Appl Microbiol Biotechnol. 2018;102:8153–71. doi: 10.1007/s00253-018-9239-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bragin EY, Shtratnikova VY, Schelkunov MI, Dovbnya DV, Donova MV. Genome-wide response on phytosterol in 9-hydroxyandrostenedione-producing strain of Mycobacterium sp. VKM Ac-1817D. BMC Biotechnol. 2019;19:39–9. doi: 10.1186/s12896-019-0533-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Penfield JS, Worrall LJ, Strynadka NC, Eltis LD. Substrate specificities and conformational flexibility of 3-ketosteroid 9alpha-hydroxylases. J Biol Chem. 2014;289:25523–36. doi: 10.1074/jbc.M114.575886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petrusma M, Hessels G, Dijkhuizen L, van der Geize R. Multiplicity of 3-Ketosteroid-9alpha-Hydroxylase enzymes in Rhodococcus rhodochrous DSM43269 for specific degradation of different classes of steroids. J Bacteriol. 2011;193:3931–40. doi: 10.1128/JB.00274-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petrusma M, Dijkhuizen L, van der Geize R. Rhodococcus rhodochrous DSM 43269 3-ketosteroid 9α-hydroxylase, a two-component iron-sulfur-containing monooxygenase with subtle steroid substrate specificity. Appl Environ Microbiol. 2009;75:5300–7. doi: 10.1128/AEM.00066-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mancilla RA, Little C, Amoroso A. Efficient bioconversion of high concentration phytosterol microdispersion to 4-androstene-3,17-dione (AD) by Mycobacterium sp. B3805. Appl Biochem Biotechnol. 2018;185:494–506. doi: 10.1007/s12010-017-2665-3. [DOI] [PubMed] [Google Scholar]
- 52.Shao M, Zhang X, Rao Z, Xu M, Yang T, Li H, Xu Z. Enhanced production of androst-1,4-diene-3,17-dione by Mycobacterium neoaurum JC-12 using three-stage fermentation strategy. PLoS ONE. 2015;10:e0137658. doi: 10.1371/journal.pone.0137658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim P-Y, Pollard DJ, Woodley JM. Substrate supply for effective biocatalysis. Biotechnol Prog. 2007;23:74–82. doi: 10.1021/bp060314b. [DOI] [PubMed] [Google Scholar]
- 54.Zhang X, Liu Y, Li H, Su L, Zhou L, Peng J, Shen R, Zhang Y, Shi J, Xu Z. Microemulsion system for Colletotrichum lini ST-1 biotransformation of dehydroepiandrosterone to 7α,15α-diOH-DHEA. Biochem Eng J. 2018;131:77–83. doi: 10.1016/j.bej.2017.12.014. [DOI] [Google Scholar]
- 55.Wang H, Song S, Peng F, Yang F, Chen T, Li X, Cheng X, He Y, Huang Y, Su Z. Whole-genome and enzymatic analyses of an androstenedione-producing Mycobacterium strain with residual phytosterol-degrading pathways. Microb Cell Fact. 2020;19:187. doi: 10.1186/s12934-020-01442-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parish T, Stoker NG. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology. 2000;146:1969–75. doi: 10.1099/00221287-146-8-1969. [DOI] [PubMed] [Google Scholar]
- 57.Petrusma M, Dijkhuizen L, van der Geize R. Structural features in the ksha terminal oxygenase protein that determine substrate preference of 3-ketosteroid 9α-hydroxylase enzymes. J Bacteriol. 2012;194:115–21. doi: 10.1128/JB.05838-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goude R, Parish T. Electroporation of Mycobacteria. In: Parish T, Brown AC, editors. Mycobacteria protocols. 2. Totowa: Humana Press; 2009. pp. 203–215. [Google Scholar]
- 59.Gygli SM, Borrell S, Trauner A, Gagneux S. Antimicrobial resistance in Mycobacterium tuberculosis: mechanistic and evolutionary perspectives. FEMS Microbiol Rev. 2017;41:354–73. doi: 10.1093/femsre/fux011. [DOI] [PubMed] [Google Scholar]
- 60.Nguyen L, Thompson CJ. Foundations of antibiotic resistance in bacterial physiology: the mycobacterial paradigm. Trends Microbiol. 2006;14:304–12. doi: 10.1016/j.tim.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 61.Borgers K, Vandewalle K, Festjens N, Callewaert N. A guide to Mycobacterium mutagenesis. FEBS J. 2019;286:3757–74. doi: 10.1111/febs.15041. [DOI] [PubMed] [Google Scholar]
- 62.Li Q, Ge F, Tan Y, Zhang G, Li W. Genome-wide transcriptome profiling of Mycobacterium smegmatis MC(2) 155 cultivated in minimal media supplemented with cholesterol, androstenedione or glycerol. Int J Mol Sci. 2016;17:689. doi: 10.3390/ijms17050689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiong LB, Liu HH, Xu LQ, Wei DZ, Wang FQ. Role identification and application of SigD in the transformation of soybean phytosterol to 9alpha-hydroxy-4-androstene-3,17-dione in Mycobacterium neoaurum. J Agric Food Chem. 2017;65:626–31. doi: 10.1021/acs.jafc.6b05314. [DOI] [PubMed] [Google Scholar]
- 64.Yao K, Wang F-Q, Zhang H-C, Wei D-Z. Identification and engineering of cholesterol oxidases involved in the initial step of sterols catabolism in Mycobacterium neoaurum. Metab Eng. 2013;15:75–87. doi: 10.1016/j.ymben.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 65.Yuan J-J, Guan Y-X, Yao S-J. Evaluation of biocompatible ionic liquids for their application in phytosterols bioconversion by Mycobacterium sp. resting cells. ACS Sustain Chem Eng. 2017;5:10702–9. doi: 10.1021/acssuschemeng.7b02681. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplementary informations include homologous recombinant sequences for knocking out targeted genes in the HGMS2 mutants, primers used for the construction of knockout and knockin vectors, antibiotic resistances of the HGMS2 strain, DNA sequence of KstD2 gene, DNA sequence of KshA51 gene, amino acid alignment of KstD211 with other KstD enzymes, amino acid alignment of KshA51 with the Ksh enzymes from the HGMS2 strain, and experimental data for PCR screening of KO and KI mutants.
Data Availability Statement
Not applicable.