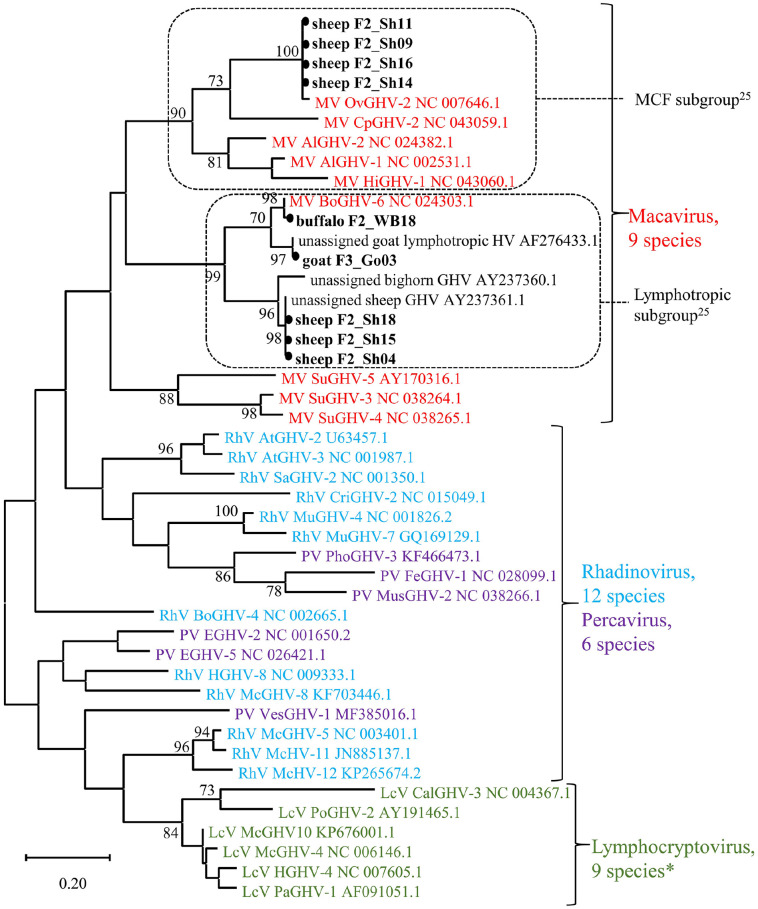
Figure 3.
Phylogenetic tree of the subfamily Gammaherpesvirinae. A maximum likelihood tree was constructed including the 9 sequences from our study as well as 1 representative of each species (n = 33) of the 4 official genera, except for 3 species in the genus Lymphocryptovirus (indicated by asterisk [*]), for which the polymerase coding region was not available. In addition, 3 unassigned viruses that by BLAST showed highest relatedness to the sheep and goat viruses were added. The 166–178-nt sequence of the polymerase coding region, flanked by the inner primer pair of the panherpesvirus nested PCR was compared. The tree with the highest likelihood is presented, and bootstrap values > 70 are provided near the respective branching point. The 4 genera are distinguished by different colors. Dashed frames indicate the subgroups within the genus Macavirus.25 The gammaherpesviruses detected in our study are highlighted with a black dot. Accessions of all reference genomes are provided after the genus and species abbreviation. The full name of the reference viruses as well as the values of the pairwise comparison are provided in Suppl. Table 3.