Figure 1.
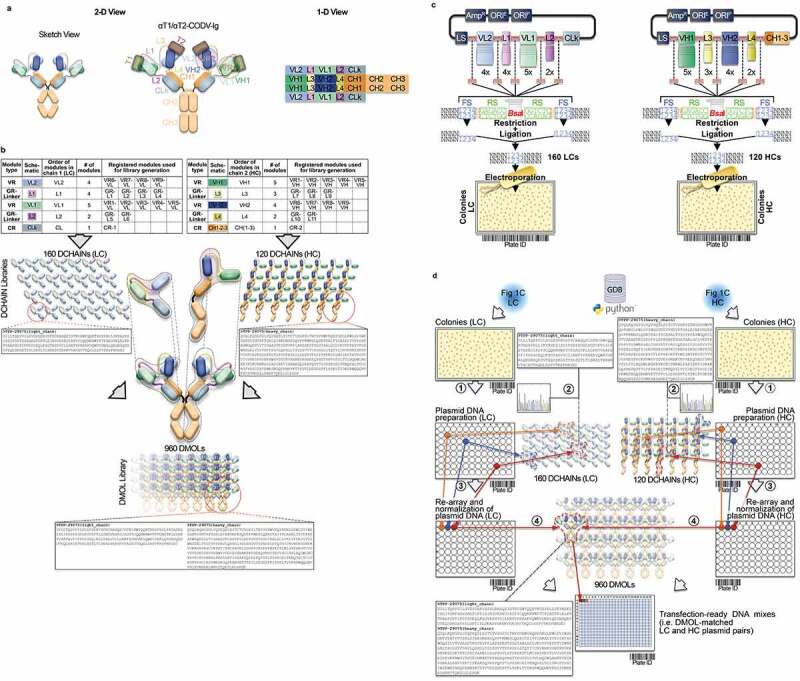
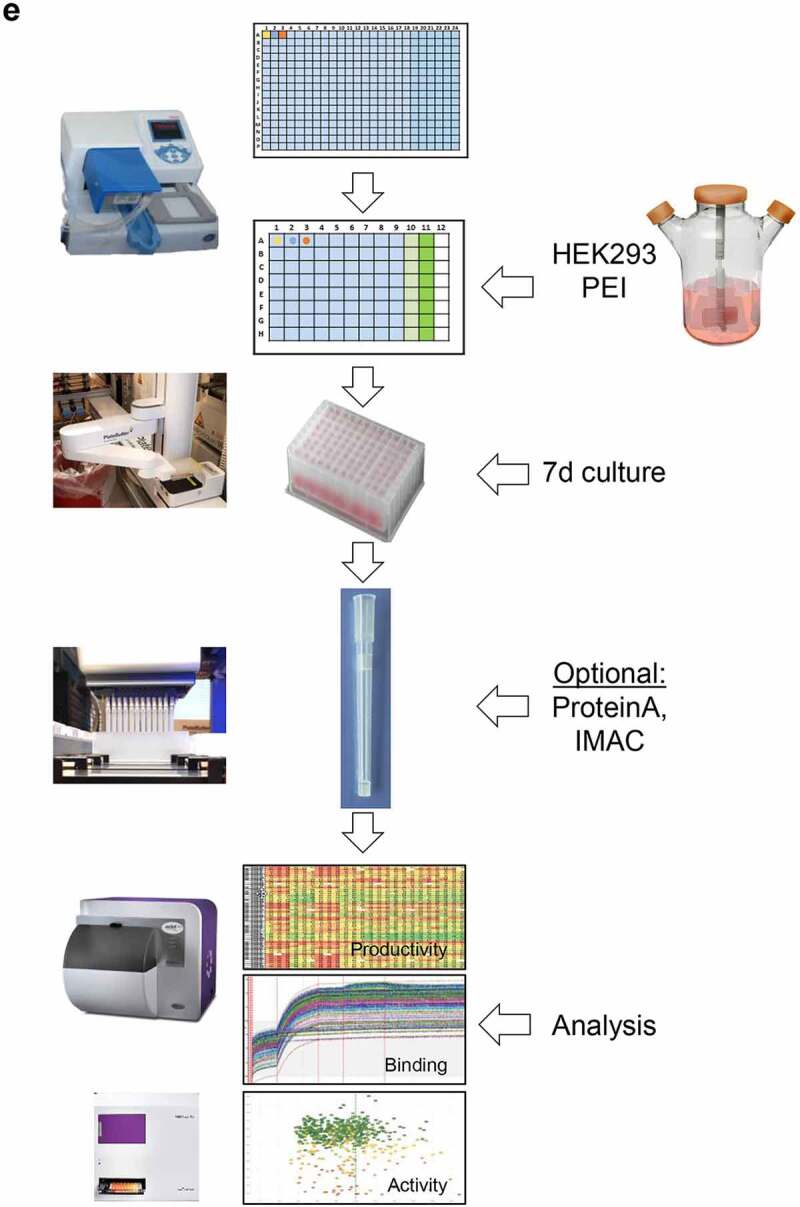
Conceptional process design for format-agnostic HT-engineering of complex MSATs. a) One-dimensional and two-dimensional views of a tetravalent, bispecific CODV molecule.19 In contrast to conventional, mono-specific immunoglobulins (IGs), in bispecific CODV-Igs one parental monoclonal antibody (mAb) here directed against target 1 (αT1; green) is extended N-terminally on both, heavy and light chains, by an additional fragment variable (Fv) domain derived from a second parental antibody directed against target 2 (α-T2; blue). However, when compared to the CODV heavy chains (HC), where the variable heavy chain domains of αT1 (VH1) are located N-terminally followed by VH2s and the heavy chain constant domains (CH; brown), the light chains (LC) carry the αT1 variable light chain domains (VL1) inserted between the VL2 domains and the constant domains of the light chains (CL; gray) resulting in a crossed-over arrangement of the variable regions (VRs) in the CODV format. Per definition, VLs and CLs are connected by peptide linkers L1 and L2, VHs and the first constant domains (CH1) of the HCs are connected by linkers L3 and L4, respectively. Full αT1- αT2-CODV-Igs are oligomerization products of two LCs and two HCs. b) In-silico MSAT reference library design by modular assembly templates. The pre-configured template for the CODV-Ig protein format includes information on the arrangement and pairing of chains, the sequential order of functional modules on chain level (i.e., variable regions (VRs), constant regions (CRs) and linker generic regions (GRs)) as well as the arrangement of functional modules between chains (e.g., cross-over pairing of VL1 with VH1 and VL2 with VH2 as depicted in a)). Pre-registered sequence building blocks can be chosen from the database (Supplementary Figure 2). As schematically shown in the idealized example five binding domains for the first CODV-Ig position (VR1 – VR5; green), four binding domains for the second CODV-Ig position (VR6-VR9; blue), one constant LC domain (CR-1), four linker variants for L1 (GR-L1 – GR-L4), two linker variants for L2 (GR-L5 – GR-L6), three linker variants for L3 (GR-L7 – GR-L9), two linker variants for L4 (GR-L10 – GR-L11), and one constant HC domain (CR-2) have been selected by the operator. The original view of the in-silico CODV-Ig design template as it appears within GDB is shown in Supplementary Figure 1b. After selection of building blocks, the system assembles the building blocks in a combinatorial fashion generating in-silico designs of the complete CODV-Ig molecules (Designed Molecules; DMOLs) according to the sequential order in the design template. 960 DMOLs covering the full combinatorial design space (here combinatorial assembly of 5 VRs x 4 VRs x 4 L1 linkers x 2 L2 linkers x 3 L3 linkers x 2 L4 linkers) are registered in the data base including their annotated sequences and information on the individual building blocks used to create respective DMOLs. In a further step the unique chain designs (DCHAINs) are derived from the DMOLs and registered in a chain specific manner (i.e., separate entries for light and heavy chains are created) holding similar descriptive information (e.g., annotated sequences and composition of building blocks) as their parental DMOLs. In our example 160 HC designs (4 x 4 × 5 x 2 × 1) and 120 LC designs (5 x 3 × 4 x 2 × 1) derived from the DMOLs are registered within GDB. By using pre-configured templates and pre-registered sequence modules, this workflow can be generically applied to any type of multi-chain MSAT format (see Supplementary Figure 1c). c) Chain library generation by modular cloning. The chain designs are realized on the DNA level using modular cloning technologies as described recently.20 Importantly, using this technology it is possible to assemble up to 10 DNA fragments in sequential, pre-defined order reflecting the in-silico design template.21 In brief, DNA building blocks that represent the pre-registered functional modules shown in b) are equipped with inversely oriented BsaI TypeIIS restriction sites. BsaI recognition sites (RS) are in green, one nucleotide spacers are indicted in orange, while the 4 base-pair cleavage/fusion sites (FS) within the DNA modules are indicated by “1234” in blue that allow the seamless and directed assembly of the 16 LC and 15 HC building blocks (i.e., the VLs, VHs, L1-4s and CRs) in one tube restriction/ligation reactions to result in pools of fully assembled 160 LCs and 120 HCs subcloned in an expression vector. The subcloned plasmid pools are transformed into E. coli and plated on omni-tray agar plates. d) Chain library deconvolution and molecule assembly on plasmid level. DNA sequences are compared against the in-silico generated reference libraries of DNA sequences for the theoretical LC and HC chain variants (2). Our Python-based algorithm identifies individual DNA constructs that match HC or LC reference designs and establishes a direct connection between the reference sequence and its physical DNA clone representative using bar-coded plate IDs and well positions of the latter. In a last step the identified clone sequences for all well positions are translated into amino acid sequences and uploaded to GDB where a connection of the physical clone variant to the corresponding DCHAIN entity is established. Identified HC and LC clone representatives (colored dots in plates) are then re-arrayed into new plates and their DNA concentrations are automatically normalized (3). In the next step our algorithm identifies corresponding HC and LC chains that match a MSAT DMOL reference and generates worklists for the liquid handling system to combine matched plasmid pairs in one cavity of a 384-well plate that will result in transfection ready DNA mixes (4). Matching HC/LC plasmid pairs are indicated by same color. e) Schematic representation of the automated LUOs covering transfection, expression, purification and multi parametrical analysis. For transient transfection plasmid pre-mixes are first transferred to 96 DWP and then combined with 1 ml of HEK293 suspension cells and PEI using a Multidrop dispensing unit. The DWPs are then closed with Duetz sandwich covers which ensure sufficient gas-exchange and prevent evaporation to result in mini-bioreactor like compartments22 (not shown). After 5–7 days incubation at 37°C the expression plates are harvested by short centrifugation. The supernatants are then transferred to new 96 DWPs and are either directly re-arrayed for various analytical and functional assays or, alternatively, first subjected to automated protein purification on the HamiltonStar LHD using PhyTips® with matched matrices (e.g., IMAC, Protein A) before further analysis. Quantification and binding data are usually obtained via label-free BLI technology using an OctetHTX device. HTRF-based data, such as from cell-based activity assays can be generated on a BMG Clariostar reader
Figure 1.
(Continued)
