Figure 2.
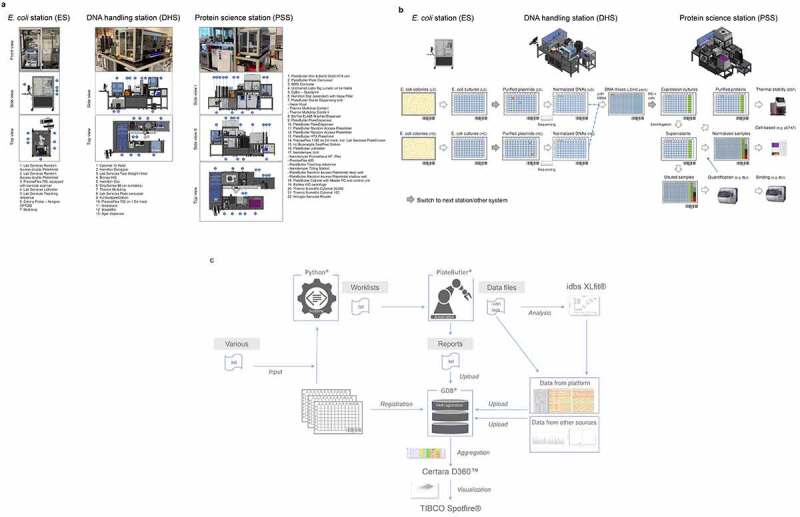
Designs of robotic platforms and automated lab unit operations. a) Robotic platforms designs. CAD drawings of the three robotic stations are shown providing side and top views including sequential numbering of integrated components. The E. coli station (ES) is designed as standalone unit to handle all E. coli-related lab unit operations (LUOs). The station is used to pick individual bacterial colonies from SBS (Society for Biomolecular Screening) compatible agar plates into 384- or 96-well plates and to inoculate 96-well-based plate formats from glycerol stocks by a cherry picking-like process using a Norgren CP7200 colony picker. Plate handling is done by a PlateButler® PreciseFlex robotic arm. A maximum of approximately 10,000 picking events can be realized per day. The fully automated DNA handling station (DHS) is designed to realize all plasmid DNA-related workflows. The station is particularly used to carry out plasmid DNA preparations using Phynexus PhyTip® chromatography technology, determine concentrations of plasmid DNAs by UV/Vis (DropSense96), re-array plasmid DNA samples and normalize the DNA concentrations of the samples in a single step, create pre-defined plasmid DNA mixtures, and generate plasmid DNA stocks for long term storage (cherry picking). To this end we integrated an automated Hamilton™ Star liquid handling device with other commercially available components for DNA concentration measurement and sample management and a PlateButler® PreciseFlex robotic arm for plate handling. The DHS accepts diverse plate and tube types which can be used with lids or seals. A maximum of 2,000 plasmid DNA preparations and 10,000 plasmid DNA normalizations can be performed per regular five-day work week. The protein science station (PSS) is a flexible robotic platform designed to conduct all cell and protein related LUOs in a fully automated fashion. The station is used to transfect mammalian HEK293 cells in 96 deep well plates in a sterile environment, to harvest protein containing cell supernatants by centrifugation and to transfer or dilute samples into other 96- or 384-well plates. Expression supernatants can be quantified by BLI- (ForteBio Octet HTX) or ELISA- (BMG CLARIOStar) based technologies. Furthermore, protein expression titers in cell supernatants can be normalized or diluted and successively applied to BLI- or ELISA-based binding assays. If required, proteins can be purified from expression supernatants using Phynexus Phytip® column technology. The purified proteins can then be quantified by UV-Vis (Unchained Labs Big Lunatic), normalized reformatted and analyzed by nanoDSF (Nanotemper Prometheus). As for the DHS, we integrated an automated Hamilton™ Star liquid handling device with other commercially available components for protein concentration measurement, analytics, and sample management and a PlateButler® PreciseFlex robotic arm for plate handling. In addition, the workstation is used to label labware with specific barcodes and to seal the samples for long term storage. In general, plates are handled with lids or seals for sterility reasons. For facility maintenance, instrument drawers and turntables are included where appropriate. To increase flexibility of the PSS, the ForteBio Octet HTX analyzer is located on a connector trolley that can be docked to the robotic station via the Lab Services PlateButler® Pier & BarQ system. This trolley can be easily disconnected and replaced by alternative instruments on further connector trolleys. Capacities of the PSS comprise over 12,000 HEK293 transfections and expression supernatant normalizations per regular five-day work week. All ES, DHS or PSS components, as listed and described in more detail in the materials and methods section and in Supplementary Table 1, are integrated and controlled by the PlateButler® software. By using a two-level principle, our ES, DHS, or PSS multi-instrument systems are set up in a distributed environment where the main application acts as the supervisor that integrates all devices. The instruments themselves are controlled by their own dedicated software which can, however, also be accessed directly by the operating scientist. All plate flows are tracked by PlateButler® software which also creates customized reports of automated experimental runs to allow for simple upload to the GDB-based laboratory information management system. b) Plateflows and automated lab unit operations. ES-operated processes: E. coli colony picking and inoculation: First, HC and LC DNA libraries are transformed into E. coli and plated onto SBS compatible omni-tray agar plates. A customized colony picking unit (Figure 2a; E. coli station (ES)) then mediates the transfer of individual colonies to 96-deep well plates (DWP) which have been supplied with 2YT growth media through an integrated multi-drop liquid dispenser. Inoculated bacterial cultures are then grown at 37°C overnight. Alternatively, glycerol stock plates are generated first from the bacterial cultures for long term storage and new 96 DWPs are inoculated from the glycerol stock plates (not shown; see materials and methods section). DHS-operated processes: DNA preparation and handling: In the next step, plasmid DNAs are extracted and purified automatically from sedimented E. coli cultures using Phynexus PhyTip® chromatography columns and subjected to DNA sequencing. Plasmid DNAs of sequence-verified clones (colored dots) are measured, normalized, and transferred to new 96-well plates. Following normalization of the plasmid DNA concentration, DNA LC/HC pairs which correspond to an in-silico MSAT DMOL reference are combined in individual cavities of 384-well plates together with oriP/EBNA vectors for stable plasmid propagation. PSS-operated processes: HEK293 cell transfection, protein expression and analysis: 384-well plates are re-arrayed to 96 DWPs that already include appropriate process and assay controls in pre-defined wells (columns 10–12). Subsequently, DNAs are mixed with HEK293F cells and PEI transfection reagent and cultivated for seven days at 37°C. After cultivation, cells are sedimented by centrifugation and cell expression supernatants are transferred to new 96 DWPs. From the new DWPs, aliquots of the samples are directly diluted into 384-well plates and first quantified by biolayer-interferometry (BLI) using an Fortebio Octet HTX instrument. Following quantification, samples are frequently normalized to appropriate concentrations before being applied to cell-based (e.g., protein phosphorylation) assays or qualitative and quantitative binding experiments using BLI- or ELISA-based technologies. For other biophysical characterization methodologies such as Nanotemper-based thermal stability assessment, samples are first immunoaffinity purified using Phynexus PhyTip® chromatography columns. c) Data processing workflow. Figure 2c describes the integrated data flow between the robotic systems (PlateButler® software), the data base (GDB®), internally developed bioinformatic tools (Python®-based) and commercial software for data analysis and visualization (idbs XLfit®, Certara D360™ and TIBCO Spotfire®). All plate and well information (including barcodes and plate hierarchies) along the screening process are consecutively fed into the data base using an a-posteriori workflow. After completion of all plate operations, the PlateButler® software generates reports as .txt files with relevant process information (e.g., barcodes of source and target plates, well to well mapping, plate types and transferred volumes) which can be uploaded into the data base (directly or after undergoing a quality control step of the pipetting operation via custom Python-based scripts). Pipetting and plate operations on the robotic systems can be initiated in different ways: 1) via pre-configured methods within the PlateButler® software (e.g., colony picking, plate copying and 96- to 384-well plate re-array operations); or 2) via a combination of pre-configured methods and worklists (e.g., hit picking, normalization, and transfection operations). Worklists are generated in .txt file format using Python-based tools and include information on source and target well positions and barcodes, as well as on plate types and volumes to be transferred. The required information for the worklist generation is automatically extracted from reports of previous plate operations or the central data repository GDB. Assay data files (including raw data) generated on the robotic system are pre-processed and analyzed via idbsXLfit® before being uploaded into the data base. Certara D360™ (Certara, Princeton, NJ) and TIBCO Spotfire® (Tibco, Palo Alto, CA) solutions are used to aggregate visualize and analyze all data generated along the value chain for the candidate molecules
