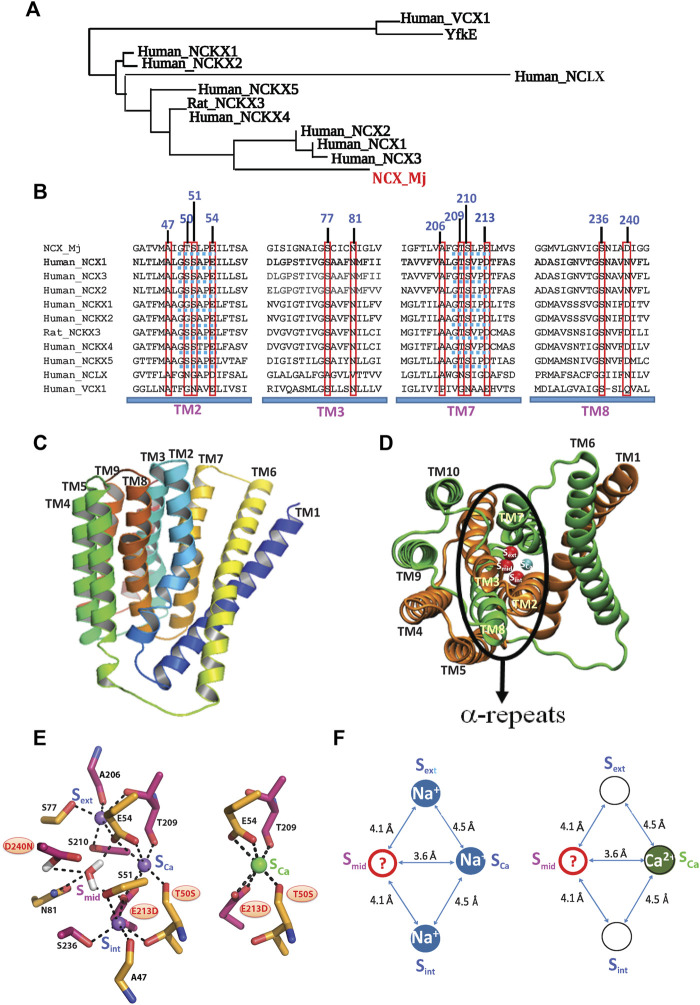
FIGURE 1.
Topological and structural highlights shared by the Ca2+/CA proteins. (A) The phylogenetically related gene families, belonging to the superfamily of the Ca2+/CA antiporters, share a common topology and structural motifs with inverted twofold symmetry, highly conserved α-repeats and ion-coordinating residues. The underlying structure-function relationships are of general interest, since the different gene products can selectively recognize and transport distinct monovalent ions (Na+, K+, H+, and/or Li+) in exchange with Ca2+. (B) Sequence alignments of the NCX, NCKX, NCLX, and CAX proteins (belonging to the superfamily of the Ca2+/CA antiporters) contain ion-coordinating residues at four transmembrane helices (TM2, TM3, TM7, and TM8). The red boxes denote the residue overlays of twelve ion-coordinating residues in the Ca2+/CA proteins (the numerations of ion coordidinating residues are assigned according to NCX_Mj). Blue dotted lines denote the helix-breaking signiture sequences (similar to GTSPLE) in different Ca2+/CA proteins. (C) The crystal structure of outward-facing (OF) NCX_Mj (PDB 3V5U) depicts ten transmembrane helices (TM1-TM10). (D) Eight helices (TM2-5 and TM7-10) form a tightly packed hub (that is perpendicularly inserted into the membrane), whereas two long and tilted helices (TM1 and TM6) are limply packed in front of a rigid eight-helix core. (E) Twelve ion-coordinating residues form four binding sites (Sint, Smid, Sext, and SCa). (F) NCX_Mj can alternatively bind either 3Na+ (at Sint, SCa, and Sext) or 1 Ca2+ (SCa). According to this model, the SCa site binds either the Na+ or Ca2+ ion, whereas the Smid site can be occupied by a water molecule (through protonated D240), but not by Na+ or Ca2+.