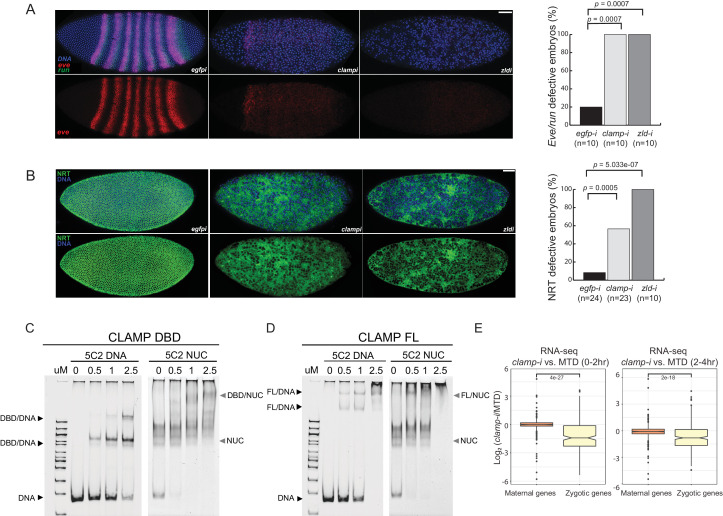
Figure 1. Novel pioneer factor CLAMP is essential for early embryonic development.
(A) Control maternal egfp depletion (left), maternal clamp depletion (middle), and maternal zld depletion (right) syncytial blastoderm stage embryos probed using smFISH for the pair-rule patterning genes run (green) and eve (red). Embryos were co-labeled with Hoechst (blue) to visualize nuclei. Scale bar represents 10 µm. Quantification (%) of eve/run defective embryos is on the right, p-values were calculated with the Fisher’s exact test; number of embryos is in parentheses. (B) Control maternal egfp depletion (left), maternal clamp depletion (middle), and maternal zld depletion (right) syncytial blastoderm stage embryos were assessed for integrity of the developing cytoskeleton using anti-NRT antibody (green) and Hoechst (blue) to label nuclei. Scale bar represents 10 µm. Quantification (%) of NRT defective embryos is on the right, p-values were calculated with the Fisher’s exact test; number of embryos is in parentheses. (C) Electrophoretic mobility shift assay (EMSA) showing the binding of increasing amounts of CLAMP DNA-binding domain (DBD) fused to MBP to 5C2 naked DNA or 5C2 in vitro reconstituted nucleosomes (Nucs). Concentrations (µM) of CLAMP DBD increase from left to right. (D) EMSA showing the binding of increasing amounts of full-length (FL) CLAMP (fused to MBP) to 5C2 DNA or 5C2 Nucs. Concentrations (µM) of CLAMP FL increase from left to right. (E) Effect of maternal clamp RNAi on maternally deposited (orange) or zygotically transcribed (yellow) gene expression log2 (clamp-i/MTD) in 0–2 hr (left) or 2–4 hr (right). Maternal versus zygotic gene categories were as defined in Lott et al., 2011. p-values of significant expression changes between maternal and zygotic genes were calculated by Mann-Whitney U-test and noted on the plot. CLAMP, chromatin-linked adaptor for male-specific lethal (MSL) proteins; smFISH, single-molecule fluorescence in situ hybridization.