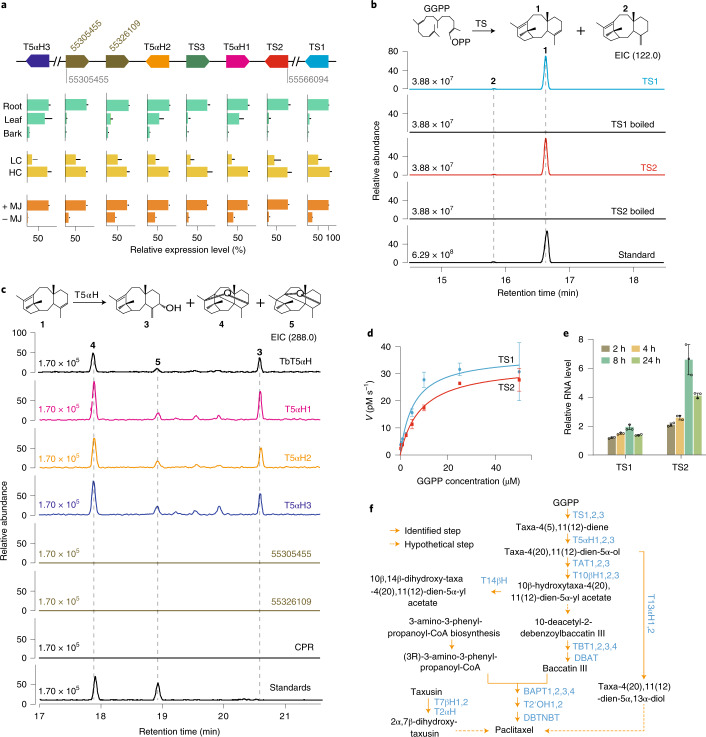
Fig. 3. Functional identification of the paclitaxel biosynthesis gene cluster.
a, Genomic architecture and expression pattern of the taxadiene cluster. The arrows indicate the relative positions and directions of the genes in the cluster. Here, 55305455 and 55566094 indicate the starting and ending positions of the cluster on pseudochromosome 9. The two unknown CYP725A genes are represented by their gene starting positions (55326109 and 55305455). TS1 and T5αH3 are located at 72105619–72109598 and 49866845–49868629 bp on chromosome 9, respectively. The relative expression levels of taxadiene cluster genes in Taxus are based on their reads per kilobase per million reads values. The expression levels of genes with high sequence similarity were distinguished on the basis of sequencing read counts of the exons that include different bases, and adjusting the alignment threshold to no mismatch. RNA-seq datasets are from roots, leaves and bark of male plants (shown in green); two T. chinensis var. mairei half-sib cell lines, HC and LC (shown in yellow); and MeJA-treated LC (+MJ) and MeJA-untreated LC (−MJ) (shown in orange). The data are shown as means ± s.d. (n = 3 biologically independent samples). b, Analysis of TS activity in vitro. The purified recombinant TS1-His and TS2-His were incubated with the substrate GGPP overnight at 32 °C. The reaction products were analysed by GC–MS. TS catalyses GGPP to produce a major product (taxa-4(5),11(12)-diene (1)) and a minor product (taxa-4(20),11(12)-diene (2)), while boiled TSs have no TS activity. m/z 122 is a characteristic ion of taxadienes. The taxadiene confirmed by NMR analysis was used as a reference standard (Standard). EIC, extracted ion chromatograms; OPP, pyrophosphoric acid. c, Analysis of the activity of T5αH and two unknown CYP725As in vitro. The in vitro enzyme assay was carried out with the purified taxadiene substrate and yeast microsomes, each including one of the six CYPs (T5αH1, T5αH2, T5αH3, TbT5αH, 55326109 or 55305455) and CPR. T5αH1/2/3 can produce three oxygenated taxadiene products (5(12)-oxa-3(11)-cyclotaxane (3), 5(11)-oxa-3(11)-cyclotaxane (4) and taxa-4(20),11(12)-dien-5α-ol (5)), whereas no catalytic compounds were observed for 55326109, 55305455 and CPR. T. brevifolia taxadiene 5-α-hydroxylase (TbT5αH), shown to have taxadiene 5-α-hydroxylase activity, was used as a positive control. d, Kinetic evaluation of GGPP oxidation catalysed by TS1 (blue circles) and TS2 (red rectangles). The x axis indicates the substrate GGPP concentration, while the y axis shows the velocity (V) of enzymatic reaction. Km = 5.5 ± 1.6 μM (TS1), Km = 8.6 ± 1.5 μM (TS2), kcat = 1705 s−1 (TS1) and kcat = 3282 s−1 (TS2). The data are shown as means ± s.d. (n = 3 biologically independent samples). e, Quantitative real-time PCR analysis of the transcription levels of TS1 and TS2 in the Taxus cell line LC treated with 100 μM MeJA for the indicated times. The relative gene expression levels are represented as the average fold change (2−ΔΔCt). The Taxus actin 1 gene (7G702435613) was used as an internal reference. The data are shown as means ± s.d. (n = 3 biologically independent samples). f, Biosynthesis pathway of paclitaxel in T. chinensis var. mairei. The solid arrows indicate the identified steps in the paclitaxel pathway, whereas the dashed arrows show the hypothetical steps. The compounds in the pathway are shown in black and the catalytic enzymes are shown in blue. T5αH, taxadiene 5-α-hydroxylase; T13αH, taxane 13-α-hydroxylase; TAT, taxadien-5-α-ol O-acetyltransferase; T10βH, taxane 10-β-hydroxylase; T14βH, taxoid 14-β-hydroxylase; T2αH, taxoid 2-α-hydroxylase; T7βH, taxoid 7-β-hydroxylase; TBT, 2-α-hydroxytaxane 2-O-benzoyltransferase; DBAT, 10-deacetylbaccatinIII 10-O-acetyltransferase; BAPT, baccatin III amino phenylpropanoyl-13-O-transferase; DBTNBT, 3′-N-debenzoyl-2′-deoxytaxol N-benzoyl transferase.