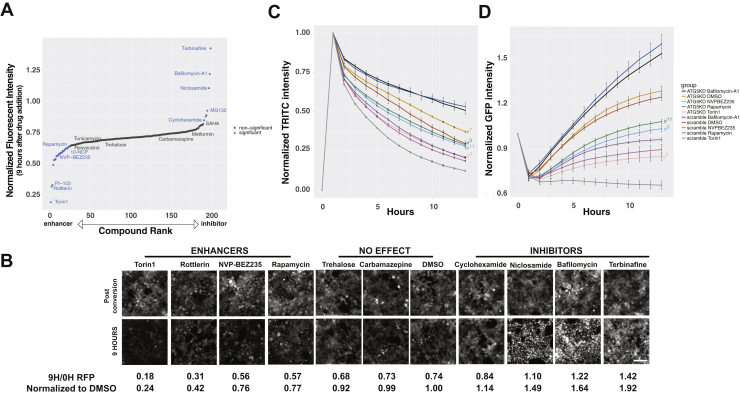
Figure 3.
Drug library screens in Dendra2-LC3 HEK293T cells confirm assay validity and identify new autophagy modulators.A, an unbiased screen of the Enzo autophagy compound library identified several known autophagy-modulating compounds, including enhancers (rapamycin, NVP-BEZ235, AKT inhibitor X) and inhibitors (bafilomycin-A1). All drugs were added at a final concentration of 10 μM via liquid handler, and autophagic flux estimated by their effects upon clearance of photoconverted (red) Dendra2-LC3 9 h after drug addition. B, representative images of red Dendra2-LC3 immediately postconversion and 9 h after drug addition, demonstrating relatively rapid clearance with application of enhancers and marked accumulation of Dendra2-LC3 with inhibitors. Both the 9 h/0 h RFP ratio and this value normalized to DMSO 9 h/0 h RFP intensity are listed below each set of images. Torin1 equivalents listed in Fig. 3-source data = [1 − (9 h/0 h RFP ratio)drug]/[1 − (9 h/0 h RFP ratio)Torin1]. Scale bar = 100 μm. C, the effect of autophagy enhancers on Dendra2-LC3 half-life is attenuated by siRNA-mediated knockdown of ATG5 2 days prior to drug application. Two-Way ANOVA indicated a significant interaction between time and treatment. F(126,300) = 15.22, p < 0.0001 and significant effects for treatment F(9,300) = 3427, p < 0.0001 and time F(14,300) = 957.3, p < 0.0001. D, similar effects are observed in the green channel. Two-Way ANOVA found a significant interaction between time and treatment F(126,300) = 20.71, p < 0.0001 as well as significant main effects for treatment F(9,300) = 721, p < 0.0001 and time F(14,300) = 320.8, p < 0.0001. In (C and D), error bars represent SEM from three biological replicates, # indicates p < 0.05 using Tukey’s multiple comparisons test with the scramble control for each drug treatment as the reference group (i.e., Scramble siRNA Torin1 versus ATG5 siRNA Torin1). Superscript number indicates the first time point when significance was achieved.