Abstract
Nephrotic syndrome (NS) is a common kidney disorder caused by dysfunction of the glomerular filtration barrier. Some genetic mutations identified in NS patients cause amino acid substitutions of kidney ankyrin repeat-containing (KANK) proteins, which are scaffold proteins that regulate actin polymerization, microtubule targeting, and cell adhesion via binding to various molecules, including the kinesin motor protein KIF21A. However, the mechanisms by which these mutations lead to NS are unclear. Here, we unexpectedly found that the eukaryotic translation initiation factor 4A1 (eIF4A1) interacts with an NS-associated KANK2 mutant (S684F) but not the wild-type protein. Biochemical and structural analyses revealed that the pathological mutation induces abnormal binding of eIF4A1 to KANK2 at the physiological KIF21A-binding site. Competitive binding assays further indicated that eIF4A1 can compete with KIF21A to interact with the S684F mutant of KANK2. In cultured mouse podocytes, this S684F mutant interfered with the KANK2/KIF21A interaction by binding to eIF4A1, and failed to rescue the focal adhesion or cell adhesion that had been reduced or morphologically changed by KANK2 knockout. These structural, biochemical, and cellular results not only provide mechanistic explanations for the podocyte defects caused by the S684F mutation, but also show how a gain-of-binding mutation can lead to a loss-of-function effect.
Keywords: KANK, eIF4A, disease mutation, gain of binding, protein-protein interaction, crystallography, FA, ankyrin-G, non-physiological binding, stress fiber
Abbreviations: co-IP, co-immunoprecipitation; eIF4A1, eukaryotic translation initiation factor 4A1; FA, focal adhesion; IDR, intrinsically disordered region; KANK, kidney ankyrin repeat-containing proteins; NS, nephrotic syndrome
Nephrotic syndrome (NS) is a kidney disorder caused by damage in glomeruli, the filtering units of kidneys, which fail to filter proteins such that proteins leak into urine, resulting in proteinuria, hypoalbuminemia, hyperlipidemia, and edema (1, 2). The glomerular filtration barrier consists of glomerular endothelial cells, the glomerular basement membrane, and highly differentiated glomerular epithelial cells, also called podocytes (3, 4). Podocytes are critical for maintaining a healthy filter barrier by preventing protein passing through the filtration barrier (3, 5, 6). By homozygosity mapping and whole-exome sequencing analysis of patients with NS, several recessive mutations in the kidney ankyrin repeat-containing (KANK) protein were identified (7).
KANK genes were first discovered as a tumor suppressor of kidney cancer, and mutation or depletion of KANK causes a variety of cancers and genetic diseases (8, 9, 10, 11, 12). Domain and phylogenetic analyses showed that the KANK protein family consists of four members (KANK1–4) in vertebrates (9). All KANK proteins are characterized by a short sequence in the N-terminus (KN motif), a coiled-coil domain, and a highly conserved C-terminal ankyrin-repeat domain (hereafter, ANKRD) (13). KANK proteins function as scaffolds linking the cytoskeletal actin filaments to integrin-mediated focal adhesions and recruiting microtubules to focal adhesion sites by interacting with various target proteins, including talin, liprin-β1, and the kinesin motor protein KIF21A (14, 15, 16, 17).
Studies have shown that knocking down KANK genes in cultured podocytes leads to abnormal podocyte motility and NS-associated mutations, including S181G and S684F in human KANK2, interfere with podocyte migration (18), indicating the important role of KANKs in maintaining normal podocyte functions. However, it remains unclear how these mutations disrupt the function of KANK proteins and consequently lead to NS.
Among the four identified missense mutations in KANK genes identified in NS patients (Fig. 1A), S684FKANK2 and Y801HKANK4 are located in the ANKRDs of KANKs. The ANKRD serves as a protein-binding module in KANKs that associates with KIF21A (15, 19). Our recent structural study on the ANKRDs of KANKs indicates that the Y801H mutant destabilizes the folding of ANKRD, whereas the S684FKANK2 mutation, corresponding to S1179F in KANK1 (Fig. 1A, Table S1), has little effect on either the ANKRD folding or the binding of ANKRD to KIF21A (20). To simplify the description, both S684FKANK2 and S1179FKANK1 are referred to as SF hereafter.
Figure 1.
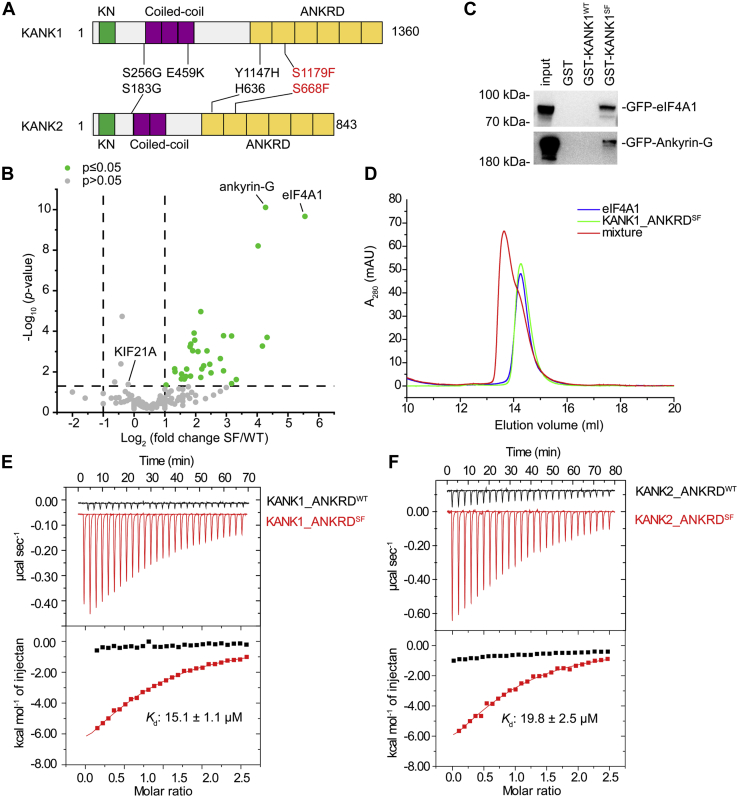
The SF mutation-induced specific interaction between KANK_ANKRD and eIF4A1.A, domain organizations of KANK1 and KANK2. Four reported genetic mutations related to NS and their corresponding mutations in KANK1 and KANK2 are indicated. The amino acid sequence numbering in is based on the mouse isoforms of KANK1 and KANK2. The uniport entry, NCBI accession number, and corresponding SF mutation numbering of different KANK proteins have been listed in Table S1. B, the volcano plot showing relative protein abundances and significant changes in mass spectra of KANK_ANKRD WT versus S1179F samples. The −log10 (p-value) is plotted against the log2 (fold change: S1179F/WT). Comparing S1179F mutant with WT samples, eIF4A1 (accession number: IF4A1_MOUSE), and ankyrin-G (accession number: ANK3_MOUSE) showed an increase of the SF/WT fold change of 47 and 19, respectively. The proteins with the increment larger than two folds were indicated in green color. C, GST pull-down assays showing co-immunoprecipitation of GFP-eIF4A1 or GFP-ankyrin-G from cell lysate using GST (lane 2), GST-KANK1_ANKRDWT (lane 3) or GST-KANK1_ANKRDSF (lane 4) as baits. D, analysis of the KANK1_ANKRDSF/eIF4A1 complex formation by analytical gel filtration chromatography using 50 μM proteins and their mixture. E, ITC measurements showing that eIF4A1 bound to KANK1_ANKRDSF but not to KANK1_ANKRDWT. F, ITC measurements showing that eIF4A1 also bound to KANK2_ANKRDSF but not to KANK2_ANKRDWT.
In this study, we were surprised to find that the SF mutation induces the nonphysiological binding of KANK proteins to a number of proteins, including the eukaryotic translation initiation factor 4A1 (eIF4A1) and the membrane-associated scaffold protein ankyrin-G. The crystal structure of KANK1_ANKRDSF in complex with eIF4A1 revealed that the SF mutation creates a new binding site for eIF4A1 and ankyrin-G, which largely overlaps with that for KIF21A. Biochemical analysis showed that the SF-induced binding of eIF4A1 to KANK proteins reduces KIF21A binding in solution and podocyte cells. We further linked the SF mutation-induced defects in podocyte adhesion and morphology to the disrupted binding of KIF21A to KANK2. Together, our structural, biochemical, and cellular findings provide novel mechanistic insights into understanding the disease-causing mutations of KANKs.
Results
Biochemical identification and characterization of partner binding induced by NS-associated KANK mutants
Considering that the serine residue corresponding to the site of SF mutation is highly conserved (Fig. S1) and located on the protein surface (20), it is likely that the serine-to-phenylalanine substitution disrupts the physiological binding of KANKs to an unknown protein and therefore interferes with podocyte functions. To identify the potential proteins with SF-mutation-sensitive binding to KANKs, we performed GST pull-down experiments coupled with mass spectrometry. Given the high sequence similarity between the ANKRD sequences of KANK1 and KANK2, we used a GST-tagged ANKRD fragment of KANK1 with or without SF mutation as bait and mouse kidney tissue lysate as prey. Consistent with our previous finding (20), SF mutation does not interfere with the binding of KANK1 to KIF21A, as indicated by the KIF21A peptides identified in the mass spectra (Fig. 1B). However, contradicting our prediction that SF mutation impairs the binding of KANK1 to certain ANKRD-binding proteins, 38 proteins were enriched in the experiment using the SF mutant with a level higher than those in the experiment using wild-type KANK1_ANKRD (Fig. 1B). Among these proteins, eukaryotic translation initiation factor 4A isoform 1 (eIF4A1) and ankyrin family member ankyrin-G exhibited the highest SF/WT fold change.
To confirm our striking finding that a single SF point mutation in ANKRD is able to induce the binding of ANKRD to eIF4A1 and ankyrin-G, we performed a co-immunoprecipitation (co-IP) experiment by overexpressing eIF4A1 or ankyrin-G with the wild-type (KANK1WT) or SF (KANK1SF) protein. Consistent with our mass spectrometry results, the co-IP assay showed that eIF4A1 and ankyrin-G interact with KANK1SF but not KANK1WT (Fig. 1C).
We further validated the SF mutation-induced interactions between eIF4A1 and KANK1_ANKRDSF or KANK2_ANKRDSF using purified proteins. In line with the aforementioned findings, an analytical gel filtration analysis indicated that eIF4A1 interacts with the SF but not wild-type ANKRDs in KANK1 and KANK2 (Fig. S2). Quantitatively, the SF proteins of KANK1 and KANK2 bind to eIF4A1 with a moderate affinity, with a Kd of 15 to 20 μM, whereas no binding was detected between eIF4A1 and wild-type ANKRD in either KANK1 or KANK2 via isothermal titration calorimetry (ITC) (Fig. 1, D and E). In a control experiment, KANK2_ANKRDSF and KANK2_ANKRDWT bound to KIF21A with comparable Kd values (Fig. S3).
SF mutation-induced binding is specific to the N-terminal domain of eIF4A1
The eIF4A family contains three members, eIF4A1-3 (21), which are ATP-dependent RNA helicases with two Rec-A-like domains, named NTD and CTD, respectively (Fig. 2A). Despite their high sequence identity (65–90%) with eIF4A1, neither eIF4A2 nor eIF4A3 shows detectable binding to KANK1_ANKRDSF (Fig. 2, B and C), indicating that the SF mutation-induced interaction between eIF4A1 and KANK proteins has isoform specificity.
Figure 2.
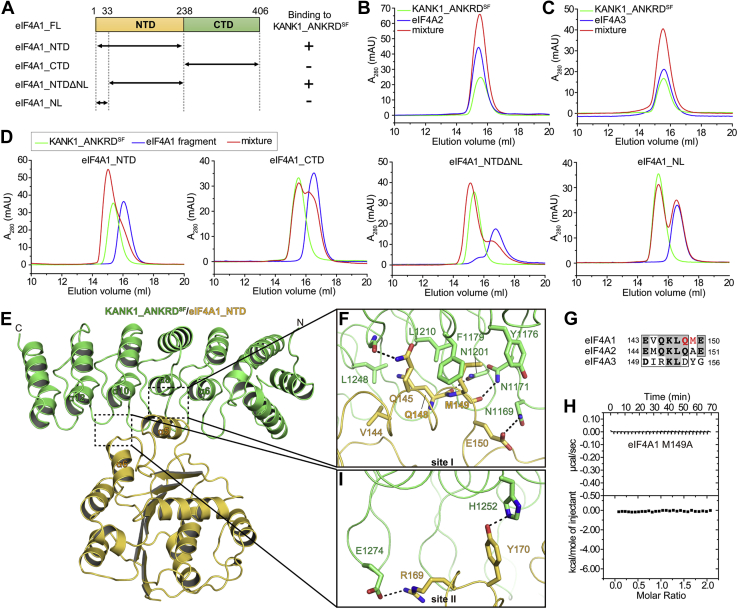
eIF4A1 bound to KANK1_ANKRDSFvia the NTD of eIF4A1.A, domains organization of eIF4A1 and constructs used in the ANKRDSF-binding region mapping. B and C, analysis of binding between eIF4A2 (B) or eIF4A3 (C) and KANK1_ANKRDSF using gel filtration chromatography. The concentrations were 40 μM for each sample in panel B and 25 μM for each sample in panel C. D, mapping the ANKRDSF-binding site in eIF4A1 by analytical gel flirtation. The protein concentration was 50 μM for each sample. E, overall structure of the KANK1_ANKRDSF/eIF4A1_NTD complex. The KANK1/eIF4A1 interface contains two sites, site I and II, indicated respectively in two dashed boxes. F and I, atomic details of the KANK1/eIF4A1 binding interface in site I (F) and II (I) were shown. Hydrogen bonds and salt bridge are indicated by dashed lines. G, sequence alignment of the α5-helices from mouse eIF4A proteins. H, ITC analysis showing no detectable binding between the M149A mutant of eIF4A1 and KANK1_ANKRDSF.
To further characterize the specific binding of eIF4A1 to the SF mutants of KANK1 and KANK2, we mapped the minimal region in eIF4A1 required for ANKRDSF binding. We purified the NTD (residues 1–237) and CTD (residues 238–406) fragments of eIF4A1 and found that only the NTD fragment interacts with KANK1_ANKRDSF (Fig. 2D). As ANKRD was found to interact with a loop region in KIF21A (15) and because eIF4A1_NTD contains a highly flexible loop in the N-terminus, we suspected that this loop may be involved in ANKRDSF binding. However, analytical gel filtration results showed that the folded region in the NTD fragment without the flexible loop is necessary and sufficient for the specific binding of eIF4A1 to ANKRDSF (Fig. 2D).
SF mutation generates a target-binding site on the ANKRD of KANK1/2
To understand how the SF mutation alters the binding property of ANKRD and leads to the abnormal recognition of eIF4A1, we determined the crystal structure of KANK1_ANKRDSF in complex with eIF4A1_NTD at 2.5 Å resolution (Table 1). Each asymmetric unit contains one KANK1_ANKRDSF:eIF4A1_NTD complex with 1:1 stoichiometry. Although carrying the SF mutation, KANK1_ANKRDSF in the complex adopts essentially the same α-solenoid fold as previously reported for the wild-type protein (20, 22, 23) (Fig. S4).
Table 1.
Statistics of data collection and structure refinement
| Data collection | |
| Space group | P3121 |
| Unit cell parameter (Å) | a = 137.628, b = 137.628, c = 57.540 |
| Resolution range (Å) | 50–2.50 (2.54–2.50) |
| No. of unique reflections | 21,919 (1716) |
| Redundancy | 10.7 (11.0) |
| I/σ | 21.3 (1.8) |
| Completeness (%) | 100 (100) |
| Rmeas (%)a | 9.0 (108.8) |
| CC1/2b | 0.99 (0.88) |
| Structure refinement | |
| Resolution range (Å) | 50–2.50 (2.61–2.50) |
| Rwork/Rfree (%)c | 19.1/21.6 (29.2/32.3) |
| R.M.S.D. bonds (Å)/angles (°) | 0.002/0.572 |
| Average B factor (Å2) | 51.31 |
| No. of atoms | 3521 |
| Proteins atoms | 3477 |
| Water atoms | 13 |
| Other solvent molecules | 31 |
| Ramachandran plot | |
| Favored/allowed/outliers (%) | 97.58/2.42/0.00 |
Values in parentheses represent the values for the highest-resolution shell.
Rmeas = Σhkl [n/(n-1)]1/2 [(Σi |Ii-<I>|)/Σi |Ii|], Ii is intensity of the ith observation for reflection hkl and <I> is its average.
CC1/2 is the correlation of one half of randomly chosen observations to the other half.
Rwork = (Σhkl ||Fobs|-|Fcalc||)/Σhkl |Fobs|, where Fobs and Fcalc are observed and calculated structure factors, respectively. Rfree = (ΣT ||Fobs|-|Fcalc||)/ΣT |Fobs|, where T is a test data set of about 5% of the total reflections randomly chosen and set aside prior to refinement.
eIF4A1_NTD binds to the concave groove formed by α6, α8, loops α5–α6, and α7–α8 of KANK1_ANKRDSF with two binding sites, sites I and II (Fig. 2E and Fig. S5, A and B). In site I, the α5-helix of eIF4A1_NTD plays a critical role in a hydrophobic interaction with the groove of KANK1_ANKRDSF. Specifically, M149eIF4A1 in the α5-helix protrudes into a hydrophobic patch formed by F1179KANK1, Y1176KANK1, and L1210KANK1 (Fig. 2F). This structural observation is completely consistent with our biochemical finding showing the SF mutation-induced binding of ANKRD to eIF4A1, as the substitution of S1179 by a phenylalanine creates a highly hydrophobic binding spot on ANKRD to hook eIF4A1. Interestingly, M149eIF4A1, which binds directly with F1179KANK1SF, is not conserved in other eIF4A proteins (Fig. 2G). As KANK1_ANKRDSF shows no binding to eIF4A2 and eIF4A3, it is likely that replacing M149eIF4A1 with the corresponding residue in eIF4A2 (A150) or eIF4A3 (Y154) disrupts the KANK1SF/eIF4A1 interaction. Indeed, the M149A mutation in eIF4A1 abolishes the binding of eIF4A1 to KANK1_ANKRDSF (Fig. 2H and Fig. S6), confirming that the hydrophobic interaction between F1179KANK1SF and M149eIF4A1 is critical for the SF mutation-induced specific binding of KANK1 to eIF4A1.
As an extension to this hydrophobic core, V144eIF4A1 and L1248KANK1 at the binding interface enhance the hydrophobic interaction, while several hydrophilic residues strengthen the interaction in site I by hydrogen bonding (Fig. 2F). Site II binding is mainly mediated by polar interactions. R169eIF4A1 forms a salt bridge with E1274KANK1, while Y170eIF4A1 interacts with H1252KANK1 via hydrogen bonding (Fig. 2I). Consistent with the two-site binding mode, we observed lower B-factors in general for residues in sites I and II of eIF4A1_NTD than for residues in other regions of eIF4A1_NTD in the crystal structure (Fig. S5C).
eIF4A1 occupies the KIF21A-binding groove in ANKRD
Previous structural studies of the KANK/KIF21A complex revealed that the KIF21A-binding surface on ANKRD is located in the concave side of the α-solenoid fold (20, 22, 23). By comparing the two complex structures of KANK1_ANKRDSF/eIF4A1_NTD and KANK1_ANKRDWT/KIF21A, we found that eIF4A1 and KIF21A bind to the same region in the vicinity of S1179 of KANK1_ANKRD, both adopting the two-site binding mode for binding to ANKRD (20) (Fig. 3A). Furthermore, the α5-helix in eIF4A1 can be aligned well to a short α-helix in a KANK-binding fragment of KIF21A, despite the low sequence similarity between these two helices (Fig. 3B).
Figure 3.
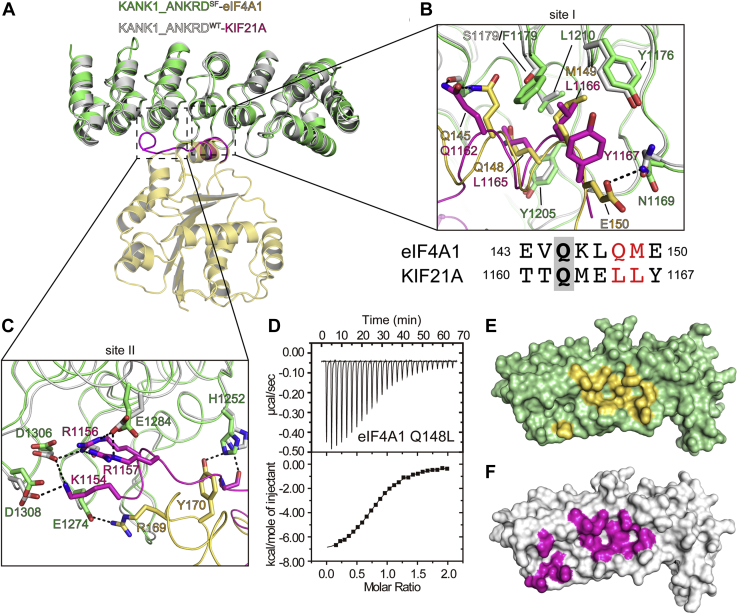
Structural comparison of ANKRD-bound eIF4A1 and KIF21A.A, structural superposition of the KANK1_ANKRDSF/eIF4A1_NTD complex and the KANK1_ANKRDWT/KIF21A complex (PDB ID: 5YAY). B and C, comparison of sites I (B) and II (C) for ANKRD binding to eIF4A1 or KIF21A. Representations use the same color code as in panel A. The ANKRD-binding sequences of eIF4A1 and KIF21A at site I were aligned guided by the overlapped structures. D, ITC analysis showing the enhanced interactions between the Q148L mutant of eIF4A1 and KANK1_ANKRDSF. The measured Kd value is 1.9 μM. E and F, surface presentations of KANK1_ANKRD with eIF4A1 bound (E) and KIF21A bound (F). Binding sites for eIF4A1 and KIF21A are highlighted in yellow and magenta, respectively.
Since eIF4A1 and KIF21A share a similar ANKRD-binding mode, why does eIF4A1 show a much weaker binding affinity than the submicromolar affinity of KIF21A (20, 22) for binding to KANK1_ANKRDSF? To address this question, we carefully analyzed the differences in the binding interfaces in the two complexes (Fig. 3, B and C). As previously reported (20, 22, 23), L1165KIF21A and L1166KIF21A of the short helix in KIF21A are critical for the binding of KIF21A to KANKs (Fig. 3C). Correspondingly, Q148eIF4A1 and M149eIF4A1 of the α5-helix are located at the same interface positions. Given the highly hydrophobic environment of the binding groove in ANKRD, the polar side chain of Q148eIF4A1 is not favorable for the interaction with KANKs, partly explaining the much lower affinity for the binding of eIF4A1 to KANK1_ANKRDSF (Fig. 3C). Consistent with our structural finding, replacing Q148eIF4A1 with leucine resulted in an eightfold higher affinity of eIF4A1 for KANK1_ANKRDSF, with a Kd of 1.9 μM (Fig. 3D).
Additionally, in site II, the extended loop of KIF21A carrying several positively charged residues, including K1154KIF21A, R1156KIF21A, and R1157KIF21A, forms salt bridges with the highly negatively charged surface of KANK1_ANKRD (20, 22, 23) (Fig. 3B). These extensive charge–charge interactions contribute to the high binding affinity of KIF21A for KANK1_ANKRD. However, eIF4A1 makes considerably less contact with KANK1_ANKRDSF at site II. Only a turn between the α5-helix and β5-strand of eIF4A1 weakly interacts with KANK1 (Fig. 3B).
SF mutation-induced binding competes with the KIF21A binding to KANK1/2
To properly perform its cellular functions, eIF4A1 forms different complexes with its physiological binding partners, such as eIF4G and PCDC4, which are required for the formation of the eIF4F translation initiation complex (24, 25) and the regulation of eIF4A1 activity (26, 27, 28, 29, 30), respectively. To analyze whether the abnormal binding of eIF4A1 to KANK1_ANKRDSF affects the physiological interaction of eIF4A1, we compared the eIF4A1 structures in complex with eIF4G, PCDC4, and KANK1_ANKRDSF. Structural superimposition shows that the binding sites for KANK1_ANKRDSF and eIF4G on eIF4A1 are distinct (Fig. S7A). The binding of KANK1_ANKRDSF to eIF4A1 may generate a mild steric clash with the second eIF4A1 molecule bound to PCDC4 in the complex containing one PCDC4 molecule and two eIF4A1 molecules (Fig. S7B). However, disruption of the second eIF4A1 binding site in PCDC4 did not impair the inhibitory effect of PCDC4 on eIF4A1 activity (29, 30). In addition, the KANK1_ANKRDSF-binding site in eIF4A1 is distal to the ATP- and RNA-binding sites (31) (Fig. S8). Considering the high abundance of the cellular eIF4A1 protein, which is required for translation, and the much lower levels of the KANK proteins in cells (8, 9, 32), the small amount of the KANK2SF protein is unlikely to interfere with the cellular function of eIF4A1.
Since the eIF4A1 and KIF21A binding surfaces overlap on KANK1 (Fig. 3, E and F), we proposed that eIF4A1 and KIF21A compete with each other for binding to KANK2SF. To test this potential interference effect of the SF mutation on the physiological binding of KIF21A to ANKRD, we performed a competition assay by using GST-fused ANKRDSF to pull down eIF4A1 in the presence of different amounts of an ANKRD-binding peptide from KIF21A (Fig. S9). Consistent with our hypothesis, the KIF21A peptide strongly prevented eIF4A1 from binding to ANKRDSF in a concentration-dependent manner. Similarly, ANKRDSF binding to ankyrin-G was also disrupted by the addition of the KIF21A peptide (Fig. S9), indicating that the binding sites for ankyrin-G and KIF21A on ANKRDSF also overlap with each other.
KIF21A binding is critical for KANK2-mediated podocyte function and morphology
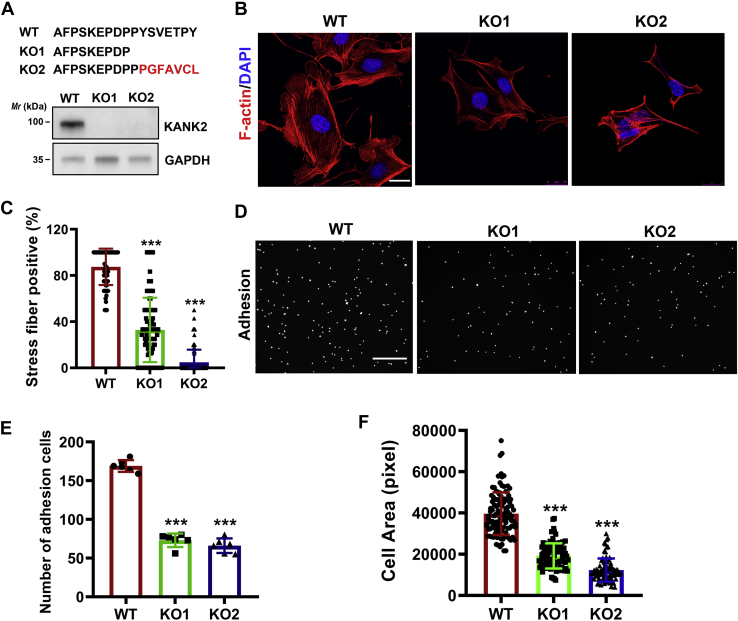
To further determine the mechanism by which the NS-associated KANK2SF mutation caused podocyte dysfunction, we first generated stable KANK2-knockout mouse podocytes (KANK2 KO1 and KANK2 KO2) using CRISPR/Cas9-mediated gene editing (Fig. 4A). As KANK proteins serve as hubs to connect the cytoskeleton and focal adhesions (16, 17), the depletion of KANK2 led to abnormal arrangement of actin filaments in both the KO1 and KO2 podocytes, as indicated by the extensive loss of stress fibers (Fig. 4, B and C). Since the cellular morphology and behavior of podocytes depend highly on the proper organization of actin filaments (6), we performed cellular assays to examine cell adhesion and morphology, which are essential for podocyte function in the formation of the glomerular filtration barrier (3, 5, 6). As expected, the cell adhesion ability and cell area of both the KANK2 KO1 and KO2 podocytes were significantly reduced compared with those of the wild-type podocytes (Fig. 4, D–F), indicating that KANK2 plays important roles in mediating podocyte function.
Figure 4.
KANK2 ablation results in dysregulation of cell adhesion and actin arrangement.A, preparation of KANK2 knockout podocytes. Protein sequence alignments showing the insertion or deletion of the amino acids in KANK2 knockout clones, resulting in early termination of KANK2 (upper panel). Immunoblotting analysis of KANK2 protein levels in wild-type mouse podocytes transfected with empty vector (WT) and in two KANK2 knockout clones (KO1 and KO2) (lower panel). B, representative images of cell morphology and immunofluorescence staining for F-actin (red) in KANK2 WT or knockout podocytes. Cell nuclei were visualized with DAPI (blue). Scale bars, 25 μm. C, quantification analysis of B. ∗∗∗p < 0.001 versus WT, n = 6 independent experiments with >112 images for each group. D, representative images for cell adhesion analysis. Each spot in the image indicates an adhesive cell. Scale bars, 800 μm. E, quantification analysis of the adhesive cells as shown in D. ∗∗∗p < 0.001 versus WT; n = 6 independent experiments. F, cell area analysis of KANK2 WT and knockout podocytes. ∗∗∗p < 0.001 versus WT, n = 5 independent experiments with 102 cells for each group.
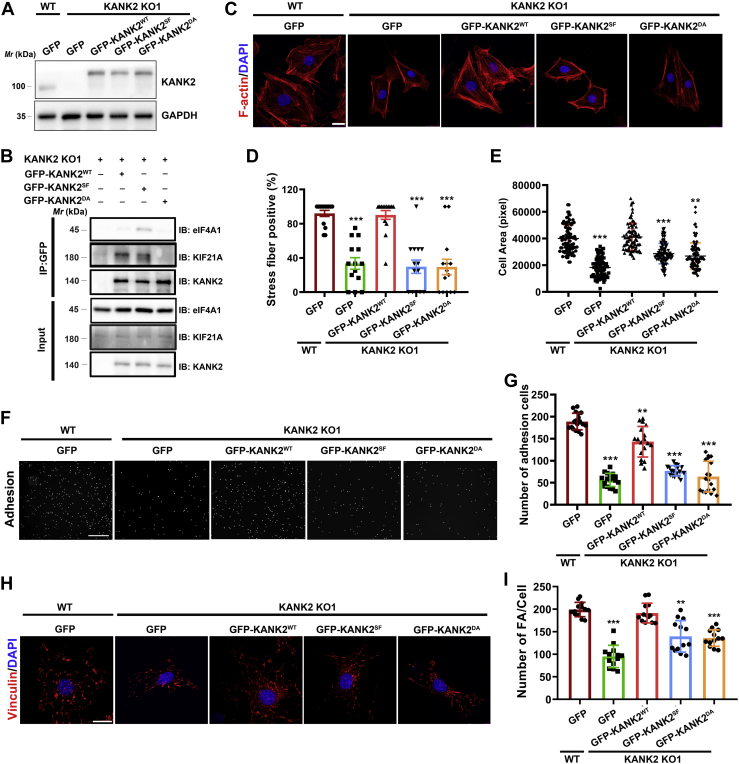
Next, we investigated whether the KANK2/KIF21A interaction contributes to KANK2-mediated podocyte function. Considering the binding differences between KIF21A and eIF4A1 to KANK1_ANKRD at site-I (Fig. 3B), we designed a DA mutation by replacing D793 and D795 with alanine, which presumably blocks the binding of KANK2 to KIF21A without diminishing the binding of KANK2SF to eIF4A1. Consistent with our design, analytical gel filtration analysis showed that KANK2DA lost the capability to bind KIF21A, while KANK2SFDA maintained SF mutation-induced binding to eIF4A1 (Fig. S10), confirming our structural findings. By using lentiviral transduction to express KANK2WT and KANK2DA in KANK2 KO1 cells at a level comparable to that of endogenous KANK2 in wild-type cells (Fig. 5A), we found that the expression of KANK2WT largely rescued the KANK ablation-induced defects in actin stress fiber formation, cell adhesion, and cell area, and the expression of KANK2DA failed to show a rescue effect (Fig. 5, C–G, GFP-KANK2DA lane). These results indicated that the KANK2/KIF21A interaction in podocytes is required for maintaining KANK2-mediated podocyte function.
Figure 5.
SF mutation in KANK2 induces podocyte defects.A, expression levels of KANK2 in the indicated cells. KANK2 WT or KO1 podocytes was infected with empty lentiviral GFP vector or lentiviral vector encoding the full-length or different mutations of KANK2. B, co-immunoprecipitation of KANK2 or its mutants with eIF4A1 and KIF21A. Cell lysates were immunoprecipitated with anti-GFP beads followed by immunoblotting with antibodies as indicated. The presence of KANK2, eIF4A1 and KIF21A in cell lysates was shown as input. C, representative images of cell morphology and immunofluorescence staining for F-actin (red) in KANK2 KO1 podocytes expressing full-length KANK2 or its mutants. Cell nuclei were visualized with DAPI (blue). Scale bars, 25 μm. D, quantification analysis of C. n = 3 independent experiments with >14 images for each group. E, cell area analysis of KANK2 KO1 podocytes expressing full-length KANK2 or its mutants. n = 5 independent experiments with >85 cells for each group. ∗∗∗p < 0.001, ∗∗p < 0.01 versus WT. F, representative images for cell adhesion analysis of KANK2 KO1 podocytes expressing full-length KANK2 or its mutants. Scale bars, 800 μm. G, quantification analysis of E. ∗∗∗p < 0.001, ∗∗p < 0.01 versus WT, n = 18. H, representative images for focal adhesion (FA) analysis of KANK2 KO1 podocytes expressing full-length KANK2 or its mutants. FAs were stained with anti-vinculin antibody. Cell nuclei were visualized with DAPI (blue). Scale bars, 25 μm. I, quantitation of FA density per cell. ∗∗∗p < 0.001, ∗∗p < 0.01 versus WT, n = 13.
SF mutation-induced binding of eIF4A1 impairs KANK2-mediated podocyte function
Consistent with our biochemical and structural findings showing that the SF mutation-induced binding of eIF4A1 inhibits KIF21A binding to KANKs, the KANK2SF mutant associates with eIF4A1 and shows a reduced KIF21A-binding capability compared with that of KANK2WT in podocytes (Fig. 5B). Because proper podocyte function requires the KANK2/KIF21A interaction, we speculated that the pathological effect of the SF mutation is due to podocyte defects caused by the reduced binding of KIF21A to KANK2. To determine whether the competition between eIF4A1 and KIF21A for KANK2SF binding impairs KANK2-regulated podocyte function, we performed a rescue experiment by expressing KANK2SF in KANK2-knockout podocytes. Similar to the outcomes observed for the KANK2DA mutant, which loses the capacity to bind KIF21A, the KANK2SF mutant showed little rescue effect on the stress fiber formation or cell adhesion that had been damaged and only weakly restored the cell area that had been decreased in the KANK2-knockout podocytes (Fig. 5, C–G, GFP-KANK2SF lane).
Given that stress fibers are focal adhesion-regulated actin cytoskeletal components and KANK2 plays an important role in regulating the focal adhesion (17), we speculate that the KANK2/KIF21A interaction might contribute to the maintenance of focal adhesion and that disrupted KIF21A binding to the KANK2 mutants causes reduced focal adhesion. Indeed, the number of focal adhesions was largely decreased in the KANK2-knockout podocytes, and the KANK2DA and KANK2SF mutants failed to restore the knockout-induced reduction of focal adhesions (Fig. 5, H and I).
As KANK2 was suggested to regulate Rho GTPase signaling in podocytes via interaction with a Rho regulator, ARHGDIA (18), it can be argued that the SF mutation interferes with KANK2 binding to ARHGDIA and thereby leads to the defects in podocyte function. However, neither the wild-type protein nor the SF mutant of KANK2_ANKRD showed detectable binding to ARHGDIA (Fig. S11). This result indicates that the SF mutation is unlikely to directly disrupt the KANK2/ARHGDIA interaction, which may be mediated by another region in KANK2. We also noted that the KANK2/ARHGDIA interaction in vivo may require posttranslational modifications; for example, SF mutation may eliminate the potential for phosphorylation. Taken together, our structural and functional data elucidated that KANK2 is vital for maintaining the normal cellular function of podocytes through its physiological binding to KIF21A, whereas the SF mutation-induced pathological binding of eIF4A1 to KANK2 reduces KIF21A binding and thereby impairs KANK2-mediated podocyte function.
Discussion
Genetic variations that lead to residue substitutions in a protein sequence, so-called missense mutations, are related to many defects and diseases. The plausible effects of missense mutations are often considered to be alterations in the protein stability, dynamics, and enzymatic activity of folded proteins or in the propensities for phase separation mediated by intrinsically disordered regions (IDRs) in proteins lacking a stable folded structure (33, 34). However, in nephrotic syndrome, the disease-associated S684F mutation in the ANKRD of KANK1/2 disrupts neither the protein folding nor the functional binding of KIF21A (20). In this study, we unexpectedly found that the S684F mutation induces the abnormal binding of eIF4A1 and ankyrin-G to KANK2 in vitro and/or in cultured podocytes. Our cellular assays further showed that SF mutation-induced binding interferes with the KANK2/KIF21A interaction and impairs KANK2-mediated podocyte functions. Thus, abnormally disruption to the physiological binding of KIF21A to KANK2 is a potential cause of the pathological consequence induced by the SF mutation in KANK2.
Although KANKs are important in mediating podocyte behaviors, the mechanistic understanding of the functions of KANKs in podocytes is very limited. By generating the KANK2 knockout podocytes, we were able to assess the potential cellular defects caused by mutations in KANK2, such as the D793A/D795A mutation in ANKRD, which specifically disrupts the binding of KIF21A to KANKs (Fig. S10) and damages actin organization in podocytes (Fig. 5C). Dysregulation of the actin cytoskeleton disrupts podocyte function, including cell adhesion and morphology (Fig. 5, E–G). Interestingly, KANKs recruit KIF21A to focal adhesions and regulate cytoskeletal dynamics (15, 16, 35). Our observations indicated that KANK2 works together with KIF21A for cytoskeletal arrangement and thus controls podocyte adhesion to the glomerular basement membrane, which is required for the proper formation and preservation of the glomerular filtration barrier. Nevertheless, the depletion of KANK2 did not completely abolish actin-related processes in mouse podocytes, suggesting that KANK1 and/or other KANKs may partially compensate for the loss of KANK2.
KANK2SF leads to podocyte defects similar to those caused by KANK2DA (Fig. 5, C–G). However, instead of directly disrupting the binding of KIF21A to ANKRD, as observed with the DA mutation, the SF mutation induces the binding of eIF4A1 and ankyrin-G to ANKRD (Fig. 1). The discovery of this gain-of-binding mutation resulting in a loss-of-function effect on KANK2 provides a potential explanation for understanding certain serine-to-phenylalanine substitutions found in other mutated proteins. Moreover, our structural analysis of the SF mutation in the KANK2SF/eIF4A1 complex indicated that single residue replacement is enough to create a binding hotspot for a new target (Fig. 2F). It is likely that proteins may undergo a similar process to gain a new function during evolution.
Compared with the Kd value measured for the KANK1/KIF21A interaction (20), the binding of eIF4A1 to the SF mutant of KANK1 or KANK2 is ~60- to 80-fold weaker (Fig. 1, E and F). How can weak eIF4A1 binding reduce strong KIF21A binding to KANK2 in podocytes? The protein concentration in the cytosol is likely a determining factor. As a translation initiation factor important for common protein expression, eIF4A1 is highly abundant in cells (36, 37). In contrast, the protein level of KIF21A in podocytes is fairly low (36, 37). Furthermore, although our structural data indicate that the KANK1SF mutant is less likely to interfere with the cellular function of eIF4A1, as discussed above, we cannot rule out the possibility that abnormal binding may interfere with the gene expression regulated by eIF4A1, thus leading to cellular dysfunction and disease progression. In addition to eIF4A1, the SF mutation induces abnormal binding of ANKRD to many other proteins, such as ankyrin-G, as shown in our mass spectrometric analysis (Fig. 1B). Hence, the KANK2/KIF21A interaction can be disrupted by the SF mutation-induced interaction occurring at the KIF21A-binding surface on KANK2.
Notably, although the ANKRDs of KANK1 and KANK2 are highly conserved, considering their variable size and the presence of different unstructured regions, the binding of eIF4A1 and other identified binding partner in this paper to the full-length proteins of KANK1SF and KANK2SF may differ from each other. Further genetic and cellular investigations are needed to confirm our findings on these KANKSF mutants.
Experimental procedures
DNA constructs and site-directed mutagenesis
DNA fragments corresponding to the ANKRD fragments of KANK1 (residues 1081–1360) and KANK2 (570–843) and the full-length proteins of eIF4A1, eIF4A2, and eIF4A3 were amplified from a mouse cDNA library (20). DNA encoding sequence of human ARHGDIA was purchased from cDNA Resource Center (Clone ID RHI0A00000). These DNA fragments were cloned into a modified pET32a vector with an N-terminal thioredoxin (Trx)-hexahistine tag. Trx-tagged proteins were used for binding assays. KANK1_ANKRD was also cloned into vector pGEX-4T-1 to generate GST-fusion proteins for GST-pulldown assays. DNA fragments of ankyrin-G were kindly provided by Dr Chao Wang (University of Science and Technology of China). The DNA fragments of full-length eIF4A1 and ankyrin-G were cloned into mammalian expression vectors containing a GFP tag for co-IP experiments. Site-directed mutagenesis was performed to create all point mutations. Mutations were confirmed by DNA sequencing.
Preparation of recombinant proteins
Recombinant proteins were produced and purified in the same way as described before (20). Briefly, BL21-CodonPlus (DE3) cells transformed with the expression plasmids were grown in LB broth to an optical density at OD600nm ~1.0 and induced with 0.1 mM isopropyl β-D-thiogalactopyranoside at 16 °C. Cells were harvested in lysis buffer (50 mM Tris-HCl pH 8.0, 500 mM NaCl, and 10 mM imidazole) and lysed using ultrahigh-pressure homogenizer (ATS, AH-BASICI). Trx-fusion proteins were purified using Ni2+-affinity chromatography and size-exclusion chromatography. GST-fusion proteins (GST-KANK1_ANKRD WT and SF mutant) were purified using glutathione sepharose and size-exclusion chromatography. Purified proteins were concentrated using Amicon Ultra-15 filter units (Millipore) in buffer containing 50 mM Tris-HCl pH 8.0, 100 mM NaCl, 1 mM EDTA, and 2 mM DTT.
Analytical size-exclusion chromatography
Analytical size-exclusion chromatography was performed using Superdex 200 Increase 10/300 GL column (GE Healthcare) with ÄKTA Pure system (GE Healthcare). Protein samples at a concentration of 25 to 60 μM were used.
GST pull-down and mass spectrometry
For GST pull-down assay, GST-KANK1_ANKRDWT and GST-KANK1_ANKRDSF (40 μg each) were incubated with 30 μl Glutathione Sepharose 4 Fastflow beads (GE healthcare), respectively. Unbound GST or GST-fusion proteins were washed with wash buffer containing 20 mM Tris pH 7.5, 100 mM NaCl, 1 mM EDTA, 1 mM EGTA, and 5% glycerol. Lysate of mouse kidney tissue was then added to the samples and incubated for 1 h with rotation. The input lysis protein amount of GST, GST-KANK1_ANKRDWT, and GST-KANK1_ANKRDSF was equal for GST pull-down. The beads were washed six times with wash buffer to remove nonspecific binding proteins. Proteins were dissociated with loading buffer and subjected to SDS-PAGE.
Proteins bound to GST-KANK1_ANKRDWT or GST-KANK1_ANKRDSF were excised from SDS-PAGE gel and trypsin digested. After desalting, the digested peptide samples were analyzed by an Easy-nLC 1000 system (Thermo Fisher Scientific) coupled with the Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific). The peptide sample was then directly loaded onto the analytical column (100 μm i.d. × 20 cm) with with 1.9 μm and 120 Å ReproSil-Pur C18 resins (Dr Maisch GmbH). The peptides were separated by a binary buffer system of 0.1% (v/v) FA in water (buffer A) and 0.1% (v/v) FA in ACN (buffer B) at a flowrate of 250 nl/min with 60 min effective gradient time and detected on the Orbitrap Fusion mass spectrometer with full MS scans performed in the Orbitrap mass analyzer over m/z range of 350 to 1550 at a mass resolution of 120,000. The MS/MS scans were implemented in the orbitrap mass analyzer with a resolution of 15,000 using an isolation window of 1.6 Da by the quadrupole mass filter. The data-dependent acquisition method in the top speed mode with cycle time of 3 s was used. The normalized collision energy of HCD fragmentation was set to 30 and the dynamic exclusion time was set to 30 s. Data were searched against Mascot5_SwissProt_mus database (9421 entries, downloaded on 2016) and analyzed using Mascot (version 1.4.1.14, Matrix Science). The maximum missed cleavages for trypsin digestion were set to 2. The methionine oxidation, asparagine, and glutamine deamidation were selected as the variable modifications, while cysteine carbamidomethylation as the fixed modification. The mass tolerances of precursor and fragment ions were set to 7 ppm and 0.02 Da, respectively. False discovery rate (FDR) of peptide spectrum matches and identified results were validated by the Percolator algorithm at 1% based on q-values. Scaffold (version Scaffold_4.6.1, Proteome Software Inc) was used to validate MS/MS-based peptide and protein identifications (Supplemental Data 1). Peptide identifications were accepted if an FDR was less than 1.0% by the Scaffold Local FDR algorithm. Protein identifications were accepted if an FDR was less than 1.0% and contained at least one identified peptide. Protein probabilities were assigned by the Protein Prophet algorithm (38). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. In total, 47 and 238 proteins were identified in the samples using GST-KANK1_ANKRDWT and GST-KANK1_ANKRDSF, respectively (Tables S2 and S3). Differences in protein abundance in different samples were compared based on the “Total Spectrum Count” of each protein as shown in Figure 1B. The mass experiment was performed once.
Binding competition assays
HEK293T cells were transiently transfected with GFP-tagged eIF4A1 or ankyrin-G. After 24 h, cells were collected and washed with PBS buffer. Resuspended cells were lysed using lysis buffer containing 50 mM Tris pH 7.5, 1% Triton X-100, 150 mM NaCl, 1 mM EGTA, 10% glycerol, 1.5 mM MgCl2, and protease inhibitor. After incubation for 30 min, cell debris was removed by centrifugation at 15,000 rpm for 20 min. Prior to adding supernatant of cell lysate to Glutathione Sepharose beads, GST-KANK1_ANKRDWT or GST-KANK1_ANKRDSF (40 μg each) was immobilized on the beads. The beads were incubated for 2 h for sufficient binding and washed with lysis buffer twice to remove unbound proteins. Then supernatant of cell lysate was added with Trx-tagged KIF21A (residues 1134–1185) at different concentrations. After incubation at 4 °C overnight, the beads were washed with lysis buffer twice and PBS buffer three times. The sample were then eluted with SDS sample buffer and loaded on SDS-PAGE gel. Proteins were detected by anti-GFP antibody (rabbit, Cell Signaling Technology) and visualized with chemiluminescence reagent (Bio-rad).
Crystallization, data collection, and structure determination
For crystallization, KANK1_ANKRD (residues 1088–1338) and eIF4A1_NTD (1–238) were mixed at a molar ratio of 1:1 and concentrated to 29.8 mg/ml. Crystals were obtained using the sitting-drop vapor diffusion method at the crystallization conditions containing 2.5 M ammonium sulfate pH 6.5 at 16 °C. Crystals of KANK1_ANKRD and eIF4A1 complex was picked up and soaked with reservoir solution containing 30% (v/v) glycerol as a cryoprotectant. X-ray diffraction data was collected at beamline BL19U1 of Shanghai Synchrotron Radiation Facility. The diffraction data were indexed, integrated, and scaled by using the HKL2000 software (39). The initial phase was determined using the molecular replacement method with the crystal structures of KANK1 (PDB code 5YAZ) and eIF4A1 (PDB code 2VSO) as search models in PHASER (40). Manual model building was carried out with Coot (41) and PHENIX (42).
Isothermal titration calorimetry (ITC)
ITC measurements were carried out using a VP-ITC MicroCal calorimeter (Malvern) at 25 °C. All protein samples were prepared in buffer of 50 mM Tris-HCl pH 7.5, 100 mM NaCl, 1 mM EDTA, and 2 mM DTT. The sample cell was filled with 20 μM eIF4A1 or its mutants and 7.0 μl aliquots of 200 μM the ANKRD protein of KANK1 or KANK2 was injected into the sample cell with 26 consecutive injections. Time interval between injections was 2 min. The first injection volume was 0.4 μl, and the observed thermal peak was excluded from the data analyses. Data fitting was performed using the program Origin7.0 with a one-site binding model.
Cell culture and establishment of KANK2 knockout stable cell lines
Conditionally immortalized mouse glomerular podocytes were propagated under permissive condition at 33 °C as previously described (43). To generate KANK2-knockout stable cell lines, we used CRISPR/Cas9-mediated gene editing. Guide RNA (gRNA) oligo designed to target the sequence of 5′- AATCGGCTCCAACCCTCGTG-3′ located at the exon 1 of KANK2, was cloned into pSpCas9n (BB)-2A-GFP (PX461 containing Cas9n was from Dr Feng Zhang, Addgene plasmid # 48140) and transfected into mouse podocytes. Single GFP-positive cells were sorted into each wells of a 96-well plate by FACS sorter (BD FACS AriaTMIII) for further propagation. Individual KANK2 knockout colonies were examined to determine disruption of the targeted locus by both DNA sequencing and immunoblotting.
Lentiviral infection
To generate KANK2 lentiviral expression vectors, cDNA encoding full-length wild-type, S668F, or D795A/D797A mutant of mouse KANK2 was cloned into GFP-tagged pLVX-IRES-Hyg vector between XhoI and BamHI sites. DNA fragments corresponding to the full-length protein of KANK2 (NP_663586) were kindly provided by Dr Reinhard Fässler. The constructed plasmids were cotransfected with psPAX2 (Addgene) and pMD2.G (Addgene) into HEK 293T cells. The culture media containing the lentivirus were collected on the third day after transfection and filtered (pore size 45 μm) and then concentrated by ultracentrifugation at 50,000g. The concentrated virus soup was immediately used or stored at −80 °C. For lentiviral infection, podocytes were cultured in growth medium until 80% confluence and then replaced with fresh medium containing lentivirus at a multiplicity of infection of 100 mixed with 8 μg/ml polybrene for 16 h. The viral infection efficiency was confirmed by immunoblotting.
Western blot analysis
Mouse immobilized podocyte cells were pooled and lysed in SDS sample buffer. Protein expression was analyzed by Western blot analysis as described previously (44). The primary antibodies used were as follows: anti-KANK1 (Proteintech), anti-KANK2 (Proteintech), anti-eIF4A1 (abcam), anti-KIF21A (purchased from ChinaPeptides Co,Ltd using a fragment containing residues NLQDGQLSDTGDLGEDIASN (45) in KIF21A to generate antibody from rabbit), anti-GAPDH (Abmart).
Co-immunoprecipitation
Cell lysates were prepared using RIPA lysis buffer (Beyotime), which included protease and phosphatase inhibitors (Roche). Samples were incubated for 30 min on ice and centrifuged at 12,000g for 15 min, 4 °C. For immunoprecipitation experiments, equal amounts of total lysates of 2 mg were incubated overnight at 4 °C with 30 μl of protein A-Sepharose beads (pre-cleaned lysates). In parallel, 30 μl of protein A-Sepharose beads was incubated for 2 h at 4 °C with the antibody of interest (2 μg) to generate the immunobeads, which were subsequently mixed with precleaned lysates and incubated overnight at 4 °C. The next day, beads were rinsed three times in TA buffer containing 20 mM Tris-HCl, pH 7.5, 5 mM sodium azide, 1 mM PMSF, and 1 mM EGTA. Proteins were eluted from Sepharose beads by adding 20 μl of 5× loading buffer containing 10% β-mercaptoethanol. Subsequently, samples were processed for Western blotting.
Cell adhesion assay
Mouse immobilized podocyte cells were trypsinized and counted by hemocytometer, 5000 cells were seeded on 24-well plates precoated with collagen type IV (10 mg/ml). After 20 min incubation at 37 °C, nonadherent cells were removed by washing with 1× PBS three times, and the adherent cells were fixed in 4% formaldehyde and then stained with 5000× Hochest. After washing, the adherent cells were determined using imaging by microscopy using a Nikon T1-SAM equipped with a ×4 objective. After five independent pictures were captured, the cell number was counted by Image J software. All experiments were repeated in triplicate for three independent experiments.
Immunofluorescence staining and cell area index
Cultured mouse podocytes were fixed with 4% paraformaldehyde for 20 min, washed three times with 1× PBS, immersed in 0.1% Triton X-100 in 1× PBS for 10 min, and then washed three times with 1× PBS again, blocked with 3% bovine serum album for 2 h, then incubated with the Alexa Fluor 555-conjugated phalloidin (Invitrogen Molecular Probes.) for 1 h at room temperature. When indicated, the cells were costained with 4,6-diamidino-2-phenylindole (DAPI) for 30 min. Images were acquired at 21 °C using a Leica TCS SP8 confocal microscope with Leica X Version:1.1.0.12420 image software. Cell areas were measured from fluorescent images in which actin was stained using the image analysis software Image-Pro Plus (Media Cybernetics). The positive of actin stress fibers was evaluated by subtracting the phalloidin-background image from the original phalloidin image.
Focal adhesion (FA) analysis
The experiments were performed as described previously (46). In brief, FAs were analyzed from fluorescent images, in which mouse podocytes were stained with anti-vinculin (Abcam) using image analysis software Image-Pro Plus (Media Cybernetics).
Statistical analysis
All experiments were repeated three or more times unless otherwise indicated. Bar graphs represent combined results from all experiments. All data represent mean ± SEM. Statistical significance was determined by two-tailed Student’s t test or one-way ANOVA. A p value less than 0.05 was considered significant.
Data availability
Atomic coordinates and structure factor amplitudes for the structure of KANK1_ANKRDSF/eIF4A_NTD have been deposited in the Protein Data Bank (https://www.wwpdb.org/) under the accession code 7DDX. All other relevant data are available from the authors.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare no competing financial interests.
Acknowledgments
We thank Dr Ting Zhang for her suggestions on the experiment design. We thank Drs Ruijun Tian, Wendong Chen, and Lin Lin for their help in MS analysis. We are grateful to Drs Anna Akhmanova, Reinhard Fässler, and Chao Wang for generously providing the plasmids encoding KANK1, KANK2, and the 190-kD isoform of ankyrin-G and Dr Chuanyue Wu for kindly providing the mouse podocyte cells. We thank the assistance of BL19U1 beamline at National Center for Protein Sciences Shanghai and Southern University of Science and Technology Core Research Facilities. This work was supported by the National Natural Science Foundation of China (Grant No. 31770791, 31870757, 31971131, 32000867, 82070728, 81900632, and 81772983), Natural Science Foundation of Guangdong Province (2016A030312016, 2020A1515011305), Department of Education of Guangdong Province (2020KZDZX1187), Shenzhen-Hong Kong Institute of Brain Science, Shenzhen Fundamental Research Institutions (2021SHIBS0002), the Shenzhen Innovation Committee of Science and Technology, China (JCYJ20190809141003834, JCYJ20200109141241950, and JCYJ20200109141212325).
Author contributions
Z. W. and C. Y. conceptualization; Y. X., C. G., W. P., C. Z., Y. D., and X. X. data curation; Y. X., C. G., and W. P. formal analysis; Z. W., Y. S., and C. Y. investigation; C. Y. project administration; Z. W., Y. S., and C. Y. supervision; Y. X. and C. G. writing—original draft; Z. W., Y. S., and C. Y. writing—review and editing.
Edited by Enrique De La Cruz
Contributor Information
Zhiyi Wei, Email: weizy@sustech.edu.cn.
Ying Sun, Email: suny@sustech.edu.cn.
Cong Yu, Email: yuc@sustech.edu.cn.
Supporting information
References
- 1.Orth S.R., Ritz E. The nephrotic syndrome. N. Engl. J. Med. 1998;338:1202–1211. doi: 10.1056/NEJM199804233381707. [DOI] [PubMed] [Google Scholar]
- 2.Eddy A.A., Symons J.M. Nephrotic syndrome in childhood. Lancet. 2003;362:629–639. doi: 10.1016/S0140-6736(03)14184-0. [DOI] [PubMed] [Google Scholar]
- 3.Scott R.P., Quaggin S.E. Review series: The cell biology of renal filtration. J. Cell Biol. 2015;209:199–210. doi: 10.1083/jcb.201410017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mundel P., Kriz W. Structure and function of podocytes: An update. Anat. Embryol. 1995;192:385–397. doi: 10.1007/BF00240371. [DOI] [PubMed] [Google Scholar]
- 5.Garg P. A review of podocyte biology. Am. J. Nephrol. 2018;47 Suppl 1:3–13. doi: 10.1159/000481633. [DOI] [PubMed] [Google Scholar]
- 6.Welsh G.I., Saleem M.A. The podocyte cytoskeleton--key to a functioning glomerulus in health and disease. Nat. Rev. Nephrol. 2011;8:14–21. doi: 10.1038/nrneph.2011.151. [DOI] [PubMed] [Google Scholar]
- 7.Gee H.Y., Saisawat P., Ashraf S., Hurd T.W., Vega-Warner V., Fang H., Beck B.B., Gribouval O., Zhou W., Diaz K.A., Natarajan S., Wiggins R.C., Lovric S., Chernin G., Schoeb D.S. ARHGDIA mutations cause nephrotic syndrome via defective RHO GTPase signaling. J. Clin. Invest. 2013;123:3243–3253. doi: 10.1172/JCI69134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarkar S., Roy B.C., Hatano N., Aoyagi T., Gohji K., Kiyama R. A novel ankyrin repeat-containing gene (Kank) located at 9p24 is a growth suppressor of renal cell carcinoma. J. Biol. Chem. 2002;277:36585–36591. doi: 10.1074/jbc.M204244200. [DOI] [PubMed] [Google Scholar]
- 9.Zhu Y., Kakinuma N., Wang Y., Kiyama R. Kank proteins: A new family of ankyrin-repeat domain-containing proteins. Biochim. Biophys. Acta. 2008;1780:128–133. doi: 10.1016/j.bbagen.2007.09.017. [DOI] [PubMed] [Google Scholar]
- 10.Guo X., Fan W., Bian X., Ma D. Upregulation of the Kank1 gene-induced brain glioma apoptosis and blockade of the cell cycle in G0/G1 phase. Int. J. Oncol. 2014;44:797–804. doi: 10.3892/ijo.2014.2247. [DOI] [PubMed] [Google Scholar]
- 11.Lerer I., Sagi M., Meiner V., Cohen T., Zlotogora J., Abeliovich D. Deletion of the ANKRD15 gene at 9p24.3 causes parent-of-origin-dependent inheritance of familial cerebral palsy. Hum. Mol. Genet. 2005;14:3911–3920. doi: 10.1093/hmg/ddi415. [DOI] [PubMed] [Google Scholar]
- 12.Ramot Y., Molho-Pessach V., Meir T., Alper-Pinus R., Siam I., Tams S., Babay S., Zlotogorski A. Mutation in KANK2, encoding a sequestering protein for steroid receptor coactivators, causes keratoderma and woolly hair. J. Med. Genet. 2014;51:388–394. doi: 10.1136/jmedgenet-2014-102346. [DOI] [PubMed] [Google Scholar]
- 13.Kakinuma N., Zhu Y., Wang Y., Roy B.C., Kiyama R. Kank proteins: Structure, functions and diseases. Cell. Mol. Life Sci. 2009;66:2651–2659. doi: 10.1007/s00018-009-0038-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen N.P., Sun Z., Fassler R. The Kank family proteins in adhesion dynamics. Curr. Opin. Cell Biol. 2018;54:130–136. doi: 10.1016/j.ceb.2018.05.015. [DOI] [PubMed] [Google Scholar]
- 15.van der Vaart B., van Riel W.E., Doodhi H., Kevenaar J.T., Katrukha E.A., Gumy L., Bouchet B.P., Grigoriev I., Spangler S.A., Yu K.L., Wulf P.S., Wu J., Lansbergen G., van Battum E.Y., Pasterkamp R.J. CFEOM1-associated kinesin KIF21A is a cortical microtubule growth inhibitor. Dev. Cell. 2013;27:145–160. doi: 10.1016/j.devcel.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 16.Bouchet B.P., Gough R.E., Ammon Y.C., van de Willige D., Post H., Jacquemet G., Altelaar A.M., Heck A.J., Goult B.T., Akhmanova A. Talin-KANK1 interaction controls the recruitment of cortical microtubule stabilizing complexes to focal adhesions. Elife. 2016;5 doi: 10.7554/eLife.18124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Z., Tseng H.Y., Tan S., Senger F., Kurzawa L., Dedden D., Mizuno N., Wasik A.A., Thery M., Dunn A.R., Fassler R. Kank2 activates talin, reduces force transduction across integrins and induces central adhesion formation. Nat. Cell Biol. 2016;18:941–953. doi: 10.1038/ncb3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gee H.Y., Zhang F., Ashraf S., Kohl S., Sadowski C.E., Vega-Warner V., Zhou W., Lovric S., Fang H., Nettleton M., Zhu J.Y., Hoefele J., Weber L.T., Podracka L., Boor A. KANK deficiency leads to podocyte dysfunction and nephrotic syndrome. J. Clin. Invest. 2015;125:2375–2384. doi: 10.1172/JCI79504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kakinuma N., Kiyama R. A major mutation of KIF21A associated with congenital fibrosis of the extraocular muscles type 1 (CFEOM1) enhances translocation of Kank1 to the membrane. Biochem. Biophys. Res. Commun. 2009;386:639–644. doi: 10.1016/j.bbrc.2009.06.109. [DOI] [PubMed] [Google Scholar]
- 20.Pan W., Sun K., Tang K., Xiao Q., Ma C., Yu C., Wei Z. Structural insights into ankyrin repeat-mediated recognition of the kinesin motor protein KIF21A by KANK1, a scaffold protein in focal adhesion. J. Biol. Chem. 2018;293:1944–1956. doi: 10.1074/jbc.M117.815779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nielsen P.J., Trachsel H. The mouse protein synthesis initiation factor 4A gene family includes two related functional genes which are differentially expressed. EMBO J. 1988;7:2097–2105. doi: 10.1002/j.1460-2075.1988.tb03049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weng Z., Shang Y., Yao D., Zhu J., Zhang R. Structural analyses of key features in the KANK1.KIF21A complex yield mechanistic insights into the cross-talk between microtubules and the cell cortex. J. Biol. Chem. 2018;293:215–225. doi: 10.1074/jbc.M117.816017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Q., Liao S., Zhu Z., Li Y., Li F., Xu C. Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins. J. Biol. Chem. 2018;293:557–566. doi: 10.1074/jbc.M117.817494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schutz P., Bumann M., Oberholzer A.E., Bieniossek C., Trachsel H., Altmann M., Baumann U. Crystal structure of the yeast eIF4A-eIF4G complex: An RNA-helicase controlled by protein-protein interactions. Proc. Natl. Acad. Sci. U. S. A. 2008;105:9564–9569. doi: 10.1073/pnas.0800418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tahara S.M., Morgan M.A., Shatkin A.J. Two forms of purified m7G-cap binding protein with different effects on capped mRNA translation in extracts of uninfected and poliovirus-infected HeLa cells. J. Biol. Chem. 1981;256:7691–7694. [PubMed] [Google Scholar]
- 26.Jansen A.P., Camalier C.E., Colburn N.H. Epidermal expression of the translation inhibitor programmed cell death 4 suppresses tumorigenesis. Cancer Res. 2005;65:6034–6041. doi: 10.1158/0008-5472.CAN-04-2119. [DOI] [PubMed] [Google Scholar]
- 27.Yang H.S., Jansen A.P., Nair R., Shibahara K., Verma A.K., Cmarik J.L., Colburn N.H. A novel transformation suppressor, Pdcd4, inhibits AP-1 transactivation but not NF-kappaB or ODC transactivation. Oncogene. 2001;20:669–676. doi: 10.1038/sj.onc.1204137. [DOI] [PubMed] [Google Scholar]
- 28.Yang H.S., Jansen A.P., Komar A.A., Zheng X., Merrick W.C., Costes S., Lockett S.J., Sonenberg N., Colburn N.H. The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol. Cell. Biol. 2003;23:26–37. doi: 10.1128/MCB.23.1.26-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loh P.G., Yang H.S., Walsh M.A., Wang Q., Wang X., Cheng Z., Liu D., Song H. Structural basis for translational inhibition by the tumour suppressor Pdcd4. EMBO J. 2009;28:274–285. doi: 10.1038/emboj.2008.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang J.H., Cho Y.H., Sohn S.Y., Choi J.M., Kim A., Kim Y.C., Jang S.K., Cho Y. Crystal structure of the eIF4A-PDCD4 complex. Proc. Natl. Acad. Sci. U. S. A. 2009;106:3148–3153. doi: 10.1073/pnas.0808275106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oberer M., Marintchev A., Wagner G. Structural basis for the enhancement of eIF4A helicase activity by eIF4G. Genes Dev. 2005;19:2212–2223. doi: 10.1101/gad.1335305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galicia-Vazquez G., Cencic R., Robert F., Agenor A.Q., Pelletier J. A cellular response linking eIF4AI activity to eIF4AII transcription. RNA. 2012;18:1373–1384. doi: 10.1261/rna.033209.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stefl S., Nishi H., Petukh M., Panchenko A.R., Alexov E. Molecular mechanisms of disease-causing missense mutations. J. Mol. Biol. 2013;425:3919–3936. doi: 10.1016/j.jmb.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsang B., Pritišanac I., Scherer S.W., Moses A.M., Forman-Kay J.D. Phase separation as a missing mechanism for interpretation of disease mutations. Cell. 2020;183:1742–1756. doi: 10.1016/j.cell.2020.11.050. [DOI] [PubMed] [Google Scholar]
- 35.Yu M., Le S., Ammon Y.C., Goult B.T., Akhmanova A., Yan J. Force-dependent regulation of talin-KANK1 complex at focal adhesions. Nano Lett. 2019;19:5982–5990. doi: 10.1021/acs.nanolett.9b01732. [DOI] [PubMed] [Google Scholar]
- 36.Schroeter C.B., Koehler S., Kann M., Schermer B., Benzing T., Brinkkoetter P.T., Rinschen M.M. Protein half-life determines expression of proteostatic networks in podocyte differentiation. FASEB J. 2018;32:4696–4713. doi: 10.1096/fj.201701307R. [DOI] [PubMed] [Google Scholar]
- 37.Rinschen M.M., Godel M., Grahammer F., Zschiedrich S., Helmstadter M., Kretz O., Zarei M., Braun D.A., Dittrich S., Pahmeyer C., Schroder P., Teetzen C., Gee H., Daouk G., Pohl M. A multi-layered quantitative in vivo expression atlas of the podocyte unravels kidney disease candidate genes. Cell Rep. 2018;23:2495–2508. doi: 10.1016/j.celrep.2018.04.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nesvizhskii A.I., Keller A., Kolker E., Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 39.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 40.Storoni L.C., McCoy A.J., Read R.J. Likelihood-enhanced fast rotation functions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 41.Emsley P., Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., McCoy A.J., Moriarty N.W., Oeffner R., Read R.J., Richardson D.C. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saleem M.A., O'Hare M.J., Reiser J., Coward R.J., Inward C.D., Farren T., Xing C.Y., Ni L., Mathieson P.W., Mundel P. A conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. J. Am. Soc. Nephrol. 2002;13:630–638. doi: 10.1681/ASN.V133630. [DOI] [PubMed] [Google Scholar]
- 44.Sun Y., Duan Y., Eisenstein A.S., Hu W., Quintana A., Lam W.K., Wang Y., Wu Z., Ravid K., Huang P. A novel mechanism of control of NFkappaB activation and inflammation involving A2B adenosine receptors. J. Cell Sci. 2012;125:4507–4517. doi: 10.1242/jcs.105023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Desai J., Velo M.P., Yamada K., Overman L.M., Engle E.C. Spatiotemporal expression pattern of KIF21A during normal embryonic development and in congenital fibrosis of the extraocular muscles type 1 (CFEOM1) Gene Expr. Patterns. 2012;12:180–188. doi: 10.1016/j.gep.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J., Liu Z., Chen K., Chen W., Fang X., Li M., Zhou X., Ding N., Lei H., Guo C., Qian T., Wang Y., Liu L., Chen Y., Zhao H. Kindlin-2 promotes rear focal adhesion disassembly and directional persistence in cell migration. J. Cell Sci. 2021;134 doi: 10.1242/jcs.244616. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factor amplitudes for the structure of KANK1_ANKRDSF/eIF4A_NTD have been deposited in the Protein Data Bank (https://www.wwpdb.org/) under the accession code 7DDX. All other relevant data are available from the authors.