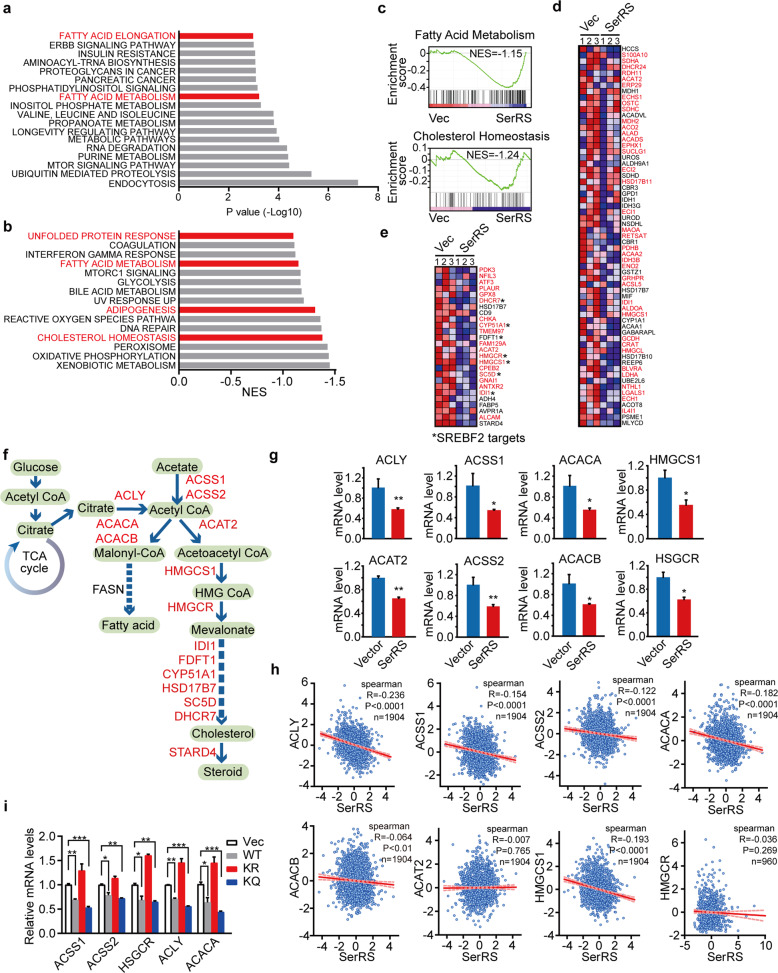
Fig. 4.
SerRS transcriptional suppresses key genes involved in the de novo lipid biosynthesis. a The top KEGG pathways that SerRS target genes identified by chromatin immunoprecipitation and DNA deep sequencing (ChIP-Seq) enrich in. b The top enriched pathways by GSEA analysis of the SerRS-suppressed genes in MDA-MB-231 cells identified by RNA sequencing. c GSEA plot of the fatty acid metabolism and cholesterol homeostasis pathways based on transcriptomes of empty vector-transfected versus SerRS-overexpressed MDA-MB-231 cells. NES, normalized enrichment score. d, e Heatmaps of the mRNA levels of enriched genes in fatty acid metabolism pathway (d) and cholesterol homeostasis pathway (e). The red signal denotes high level and the blue signal denotes low level in SerRS-overexpressed MDA-MB-231 cells. The genes in red were also SerRS target genes identified by ChIP-Seq. f The de novo lipid synthesis pathways and the key enzymes. g The mRNA levels of indicated genes in SerRS-overexpressed and empty vector-transfected MDA-MB-231 cells were further quantified by qRT-PCR (means ± SEM, n = 3, Student’s t-test, *P < 0.05, **P < 0.01). h The correlation of mRNA levels of SerRS and the indicated key genes in human breast cancers collected from TCGA database. i The mRNA levels of indicated genes in MDA-MB-231 cells transfected with empty vector (Vec), wild type or mutant SerRS were quantified by qRT-PCR (means ± SEM, n = 3, Student’s t-test, *P < 0.05, **P < 0.01, ***P < 0.001)