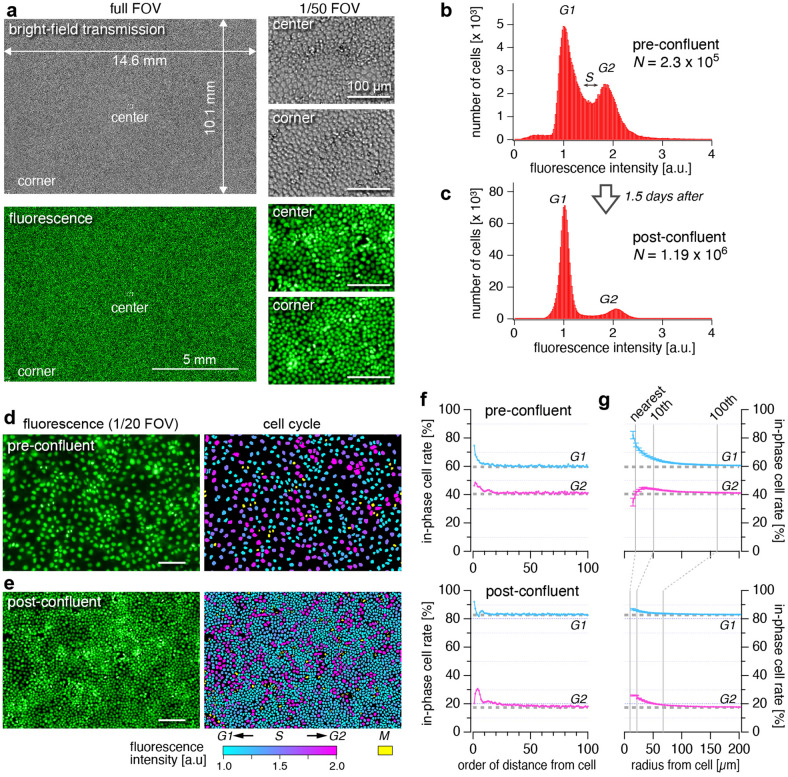
Figure 3.
Single shot detection and analysis of more than one million cells. Imaging of the cultured MDCK cells that were fixed with paraformaldehyde and stained with NucleoSeeing was performed with an excitation LED wavelength of 470 nm. (a) Full FOV image and closeup images of the area covering 1/50 of the FOV region at the center and corner, obtained with the bright-field transmission (top three) and fluorescence (bottom three) modes. (b,c) Histograms of fluorescence intensity of cells with two different cell densities (b: pre-confluent, c: post-confluent), which correspond to the populations of cells in the G1, S and G2 phases. The fluorescence intensity was normalized by the peak position for the G1 phase. (d,e) Closeup views of areas comprising 1/20 of the FOV region obtained at the pre-confluent condition (d) and the post-confluent condition (e). In the right panels, nuclei of interphase cells are painted with colors indicating fluorescence intensities of the cells, with each color corresponding to different phases of the cell cycle, i.e., cyan and magenta represent the G1 and G2 phases, respectively. Nuclei of the M phase cells are painted with yellow. Scale bars: 100 µm. (f,g) Spatial analysis of the cells in the two phases. (f) In-phase cell rate of neighboring cells as a function of order of cell-cell distance (G1: cyan, G2: magenta). (g) In-phase cell rate among the cells in the neighboring area was plotted as a function of the radius of the area (G1: cyan, G2: magenta). The error bars represent standard errors. The dashed gray lines in f and g represent the expected values for random distribution.