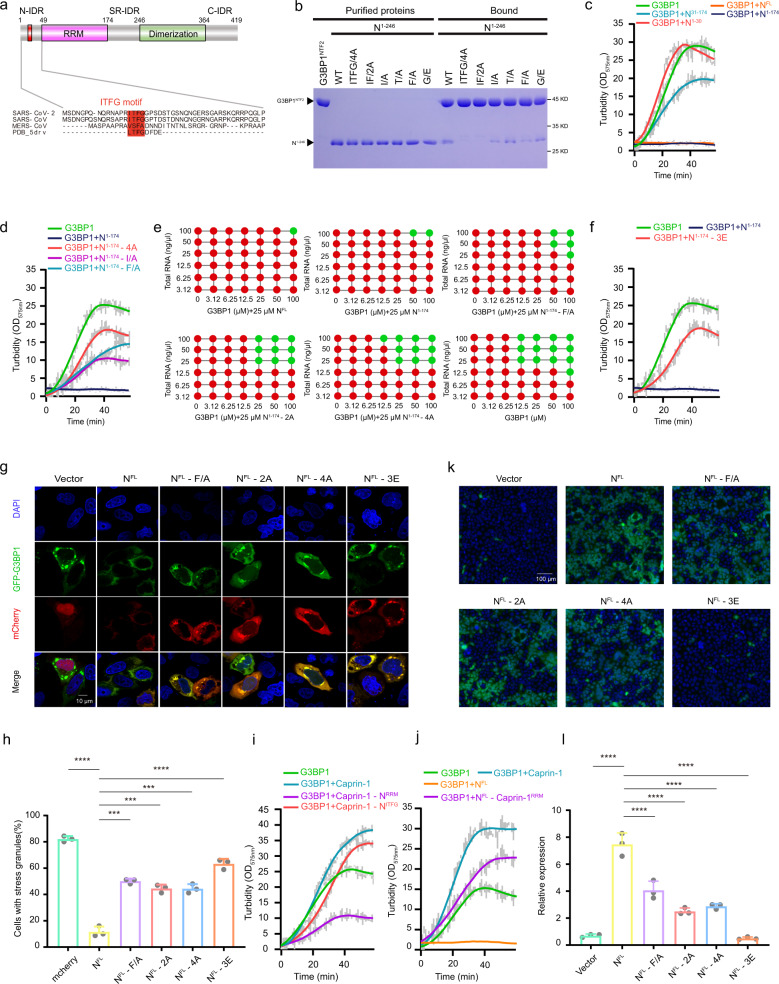
Fig. 1. SARS-CoV-2 N protein utilizes two components, the ITFG motif and RRM, to regulate G3BP1 phase separation.
a Schematic diagram of SARS-CoV-2 N protein highlights an ITFG motif located in the N-IDR. IDR intrinsically disordered region. b GST–G3BP1NTF2 pull-down of purified SARS-CoV-2 N1–246 and ITFG mutants. c SARS-CoV-2 N1–174, but not N1–30 or N30–174, is sufficient to inhibit phase separation of G3BP1 in the presence of RNA. Equimolar N fragment was mixed with MBP-G3BP1. TEV was added in solution to cleave the MBP fusion, and OD575nm was recorded over time. Data represent mean±s.d. from 3 independent experiments. d Mutations in the ITFG motif of N protein impaired its ability to inhibit G3BP1-mediated phase separation. Experiments were performed similar to (c). e Summary of the phase separation behaviors of purified recombinant G3BP1 and RNA, in the absence of N protein or the presence of various N mutants. Red: no droplet; green: droplet. f Mutation of three basic amino acids (R92E/R107E/R149E, 3E) in the RRM domain of SARS-CoV-2 N protein significantly impaired the inhibitory effect of N protein on G3BP1 phase separation. g, h Overexpression of SARS-CoV-2 N protein inhibited the formation of SGs in cells, and mutation of the ITFG motif or RRM significantly abrogated the inhibition. Data shown in (h) represent mean ± s.d. from three independent experiments. Results were evaluated by one-way ANOVA test (**P < 0.01; ***P < 0.001; ****P < 0.0001; NS, not significant). Scale bar: 10 μm. i Turbidity of G3BP1 with Caprin-1 or its mutants over time in the presence of RNA. Data represent mean ± s.d. from three independent experiments. j Turbidity of G3BP1 with Caprin-1 or N protein over time in the presence of RNA. Data represent mean ± s.d. from three independent experiments. k, l Caco-2 cells expressing N wild-type or mutants were infected with SARS-CoV-2 GFP/ΔN trVLP, and cell culture supernatant was collected to infect Caco-2-N wild-type cells. GFP signal was observed using microscopy, and cellular RNA was extracted for RT-qPCR analysis to determine viral subgenomic RNA levels. Data represent mean ± s.d. from three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Significance assessed by one-way ANOVA. Scale bar: 100 μm.